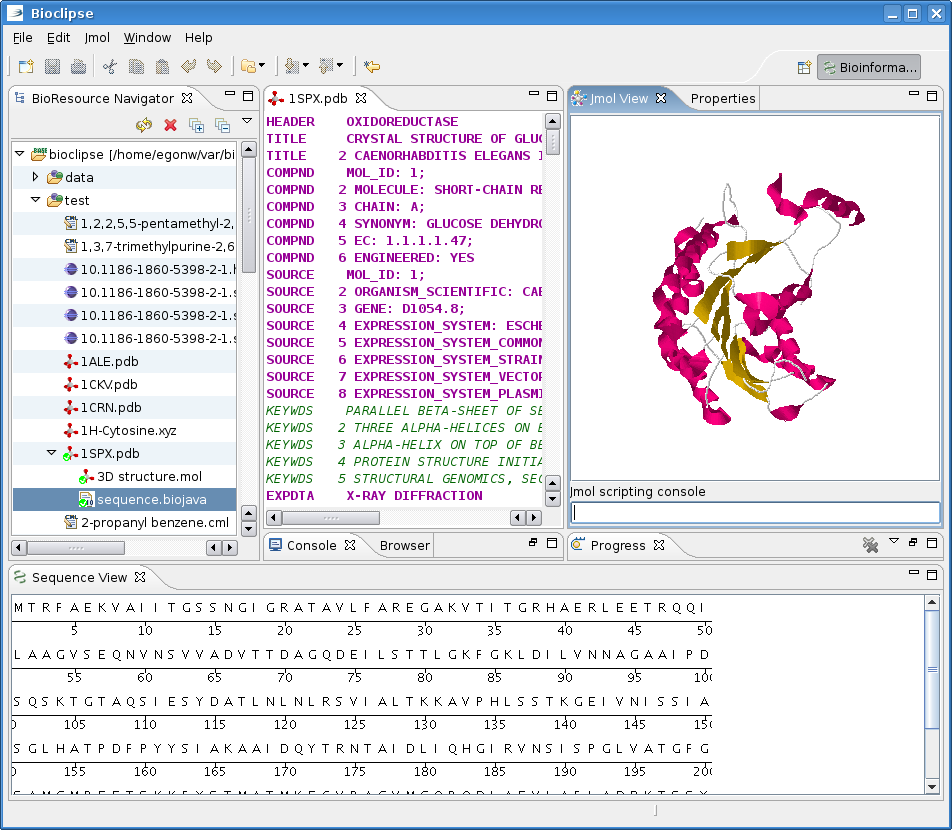
Some time ago I blogged about the ChildResourceCreator extension point in Bioclipse and hinted as using that for PDB files. which contain 3D molecular models, sequences and bibliographic information.

Some time ago I blogged about the ChildResourceCreator extension point in Bioclipse and hinted as using that for PDB files. which contain 3D molecular models, sequences and bibliographic information.
Bernard Munos at Eli Lilly & Co. wrote up a lengthy analysis on open source in drug discovery in Nature Reviews Drug Discovery: Can open-source R&D reinvigorate drug research?
Martin Szugat reported that a beta for BioJava 1.5 has been released. New features include: a new biojavax package with extension on the basic functionlity, such as the RichSequence.IOTools and the RichSequence object; a genetic algorithm library; features that allow manipulation of 3D structure files and objects; and non-HMM implementations of the NW and SW alignment algorithms.
Chemical Archeology (see Christoph’s comment) is the process of extracting chemical information from old journal articles. Some time ago, Peter Corbett from the group of Peter Murray-Rust visited the CUBIC to talk to us about Oscar3 which can do just that. That day, we already hooked OPSIN into Bioclipse . Oscar3, however, is capable of more then the name2structure of OPSIN (see also 10.1039/b411033a;
ケムインフォマティクスに虚空投げ runs a story on how to calculate geometrical properties of a 3D structure using CDK’s ForceFieldTools. This class contains a few methods to calculate distances between atoms and angles between bonds. This tools class is special as it uses vecmath GVector objects, which just contain atomic coordinates, likely suitable for extensive computation, as expected in CDK’s force field implementation.
R News just released a special issue on the use of the versatile statistics program R in chemistry. It features six articles amongst which one by Rajarshi Guha on the CDK-R bridge, and one by my supervisor and me on the use of self-organizing maps to cluster crystal structures.
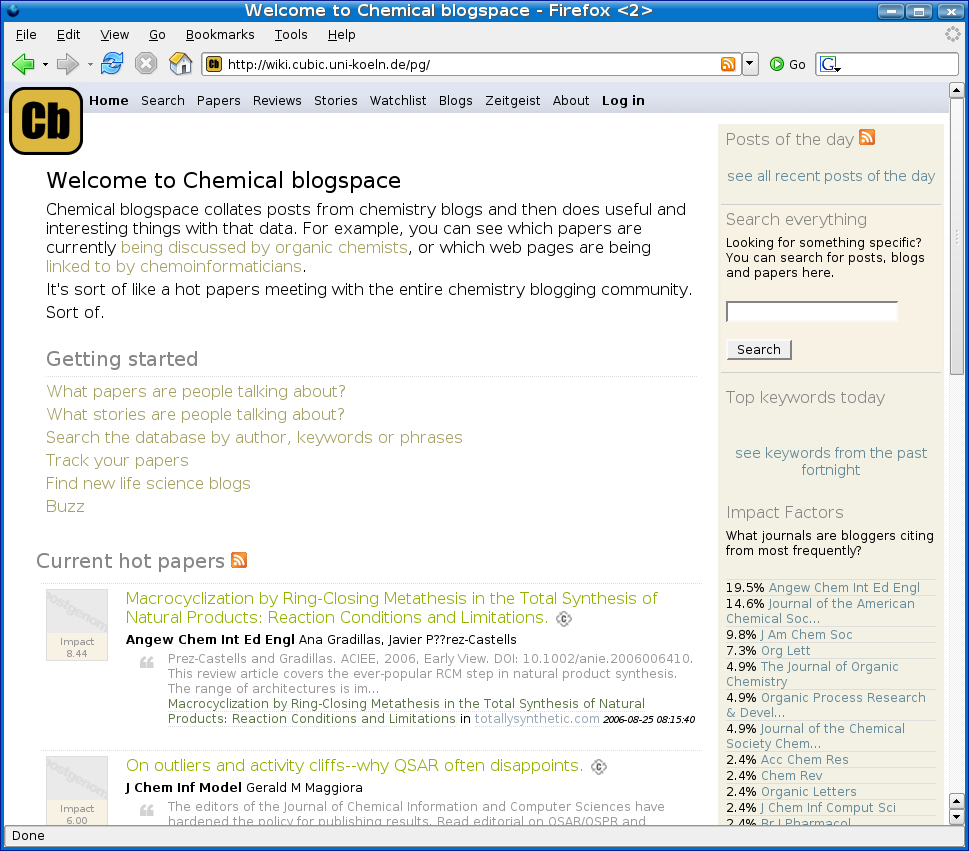
We all know chemical space; Chemical blogspace (Cb) is different: it is the chemistry discussed in blogspace. Cb is build on the opensource software of Postgenomic.com which I bloged on before. The now running Cb aggregates 19 blogs and, like the original, extracts linked (cited or reviewed) articles from literature. The system is beta, but I am happy about it already that I mention it now.
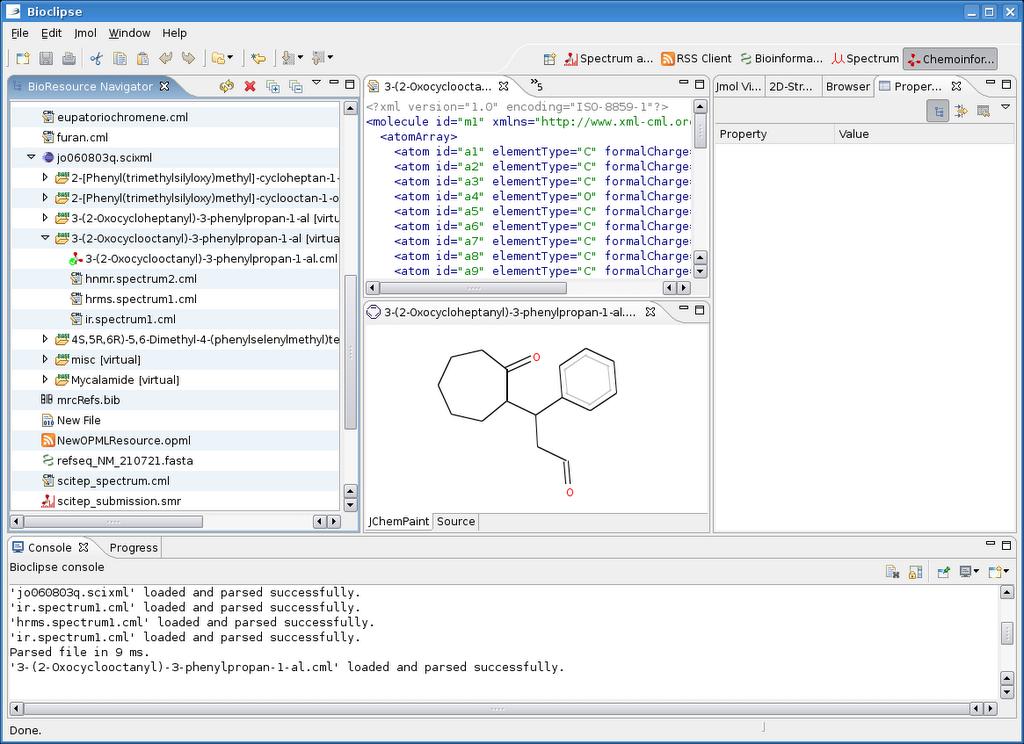
I hacked in a new extension point for Bioclipse yesterday, based on a proposal I made earlier. The new extension point (EP) is called ChildResourceCreator and allows creating child resources for a given IBioResource. One application where this is very useful is the CMLRSS application (earlier blog), or any RSS or Atom enriched with any other XML language.
Recently, a new generation of Chemical Markup Language CML users seem to hit the learning-curve-wall; there seems to be a niche in explaining the use of CML, so here goes. My new (third) blog will discuss frequently and less frequently asked questions about the use of CML.
Trepalin et al. published in Molecules the article A Java Chemical Structure Editor Supporting the Modular Chemical Descriptor Language (MCDL) (open access PDF). The applet is about 250kB (though the article mentions 200kB) in size and downloadable from the MCDL project on SourceForge (license: Public Domain). The article compares the applet with the JChemPaint applet and notes that their applet is much smaller.