
As part of a project exploring GBIF data I've been playing with displaying GBIF data on Google Maps.

As part of a project exploring GBIF data I've been playing with displaying GBIF data on Google Maps.

A quick note to myself to document a problem with the GBIF classification of liverworts (I've created issue POR-1879 for this). While building a new tool to browse GBIF data I ran into a problem that the taxon "Jungermanniales" popped up in two different places in the GBIF classification, which broke a graphical display widget I was using.

There is a great post by Jeni Tennison on the Open Data Institute blog entitled Five Stages of Data Grief. It resonates so much with my experience working with biodiversity data (such as building BioNames, or exploring data errors in GBIF) that I've decide to reproduce it here.

I gave a remote presentation at a proiBioSphere workshop this morning. The slides are below (to try and make it a bit more engaging than a desk of Powerpoints I played around with Prezi). There is a version on Vimeo that has audio as well. I sketched out the biodiversity "knowledge graph", then talked about how mark-up relates to this, finishing with a few questions.

Scott Federhen told me about a nice new feature in GenBank that he's described in a piece for NCBI News. The NCBI taxonomy database now shows a its of type material (where known), and the GenBank sequence database "knows: about types. Here's the summary: You can query for sequences from type using the query "sequence from type"[filter]. This could lead to some nice automated tools.

VertNet has announced that they have implemented issue tracking using GitHub. This is a really interesting development, as figuring out how to capture and make use of annotations in biodiversity databases is a problem that's attracting a lot of attention.
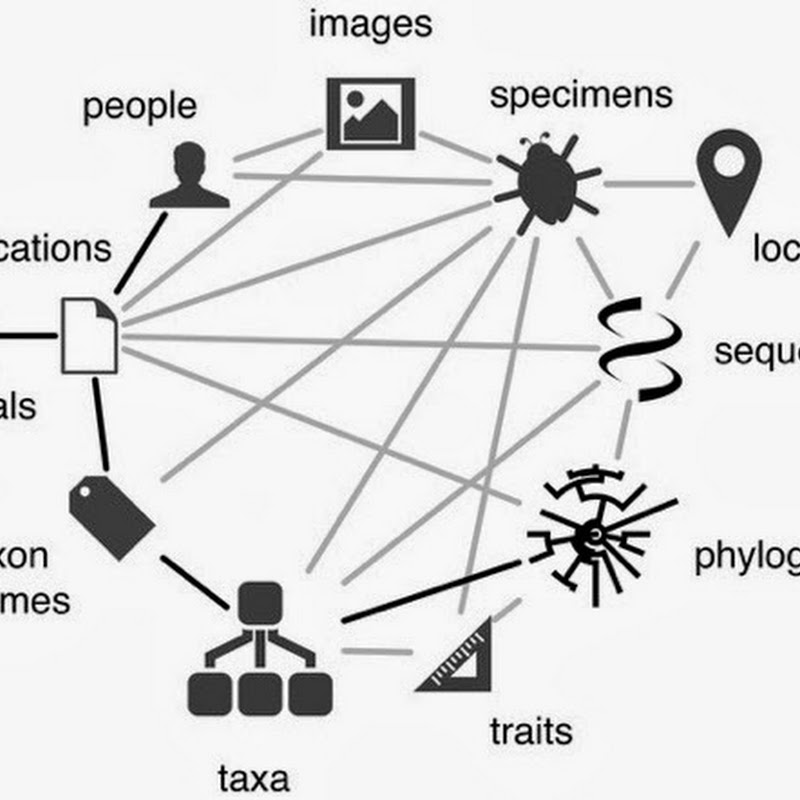
More for my own benefit than anything else I've decided to list some of the things I plan to work on this year. If nothing else, it may make sobering reading this time next year. A knowledge graph for biodiversity Google's introduction of the "knowledge graph" gives us a happy phrase to use when talking about linking stuff together.
Given that it's the start of a new year, and I have a short window before teaching kicks off in earnest (and I have to revise my phyloinformatics course) I'm playing with a few GBIF-related ideas. One topic which comes up a lot is annotating and correcting errors. There has been some work in this area [1][2] bit it strikes me as somewhat complicated. I'm wondering whether we couldn't try and keep things simple.

The following is a guest blog post by David Schindel and colleagues and is a response to the paper by Antonio Marques et al. in Science doi:10.1126/science.341.6152.1341-a. Marques, Maronna and Collins (1) rightly call on the biodiversity research community to include latitude/longitude data in database and published records of natural history specimens.

A while ago I posted BHL to PDF workflow which was a sketch of a work flow to generate clean, searchable PDFs from Biodiversity Heritage Library (BHL) content: I've made some progress on putting this together, as well as expanded the goal somewhat. In fact, there are several goals: BioStor articles need to be archived somewhere.