
Ideas on measuring the "impact" of a natural history collection have been bubbling along, as reflected in recent comments on iPhylo, and some offline discussions I've been having with David Blackburn and Alan Resetar.

Ideas on measuring the "impact" of a natural history collection have been bubbling along, as reflected in recent comments on iPhylo, and some offline discussions I've been having with David Blackburn and Alan Resetar.

Bob Mesibov (who has been a guest author on this blog) recently published a paper on data quality in in ZooKeys : In this paper Bob documents some significant discrepancies between data in his Millipedes of Australia (MoA) database and the equivalent data in the Atlas of Living Australia and GBIF (disclosure, I was a reviewer of the paper, and also sit on GBIF's science committee). This paper spawned a thread on TAXACOM, and also came
Things are finally coming together, at least enough to have a functioning demo. It looks awful, but shows the main things I want BioNames to do. One thing I'm most concerned about at this stage is the possible confusion users might experience between taxon names and concepts.

Over on Google Plus (yeah, me neither) Donat Agosti is giving me a hard time regarding the quality of some data that I am using. I've responded to Donat directly, but here I just want to quickly outline two different approaches to cleaning and reconciling bibliographic metadata.

This seems to be the season for big, arm-wavy documents about the future of biodiversity informatics (see A decadal view of biodiversity informatics: challenges and priorities). An equivalent document is being drafted based on the Global Biodiversity Informatics Conference (GBIC 2012) conference.

In an earlier post I discussed using Open Refine (formerly Google Refine) to clean and reconcile taxon names. I've added an additional service that can be used to reconcile author names that uses the Virtual International Authority File (VIAF) API.

BMC Ecology has published Alex Hardisty and Dave Roberts' white paper on biodiversity informatics: Here are their 12 recommendations (with some comments of my own): Open Data, should be normal practice and should embody the principles of being accessible, assessable, intelligible and usable.
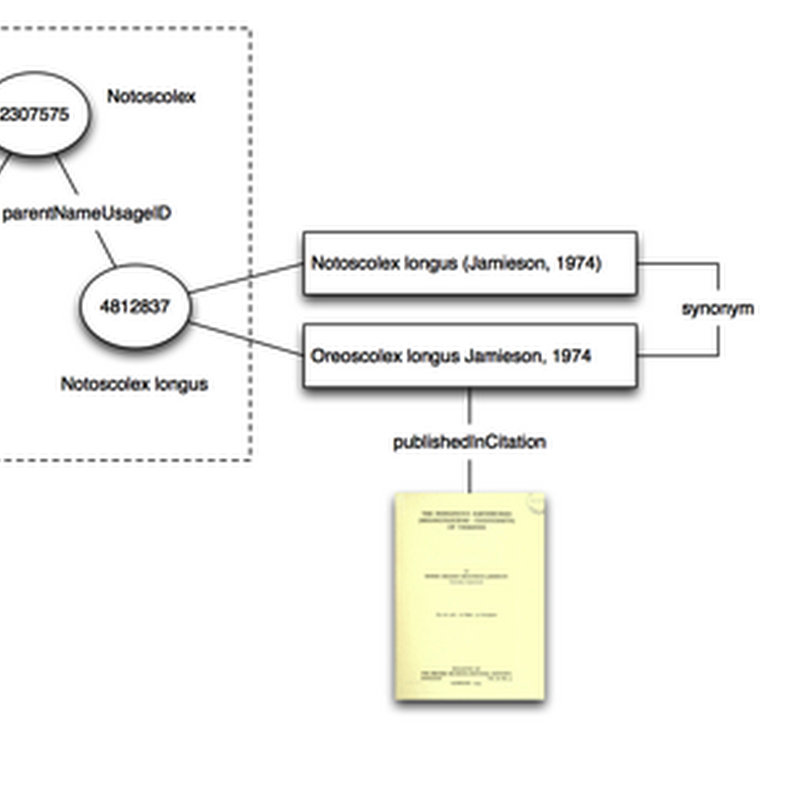
Quick notes on "taxon concepts". In order to navigate through taxon names I plan to have at least one taxonomic classification in BioNames. GBIF makes the most sense at this stage. The model I'm adopting is that the classification is a graph where nodes have the id used by the external database (in this case GBIF). Each node has one or more names attached, and where possible the names are linked to the original description.
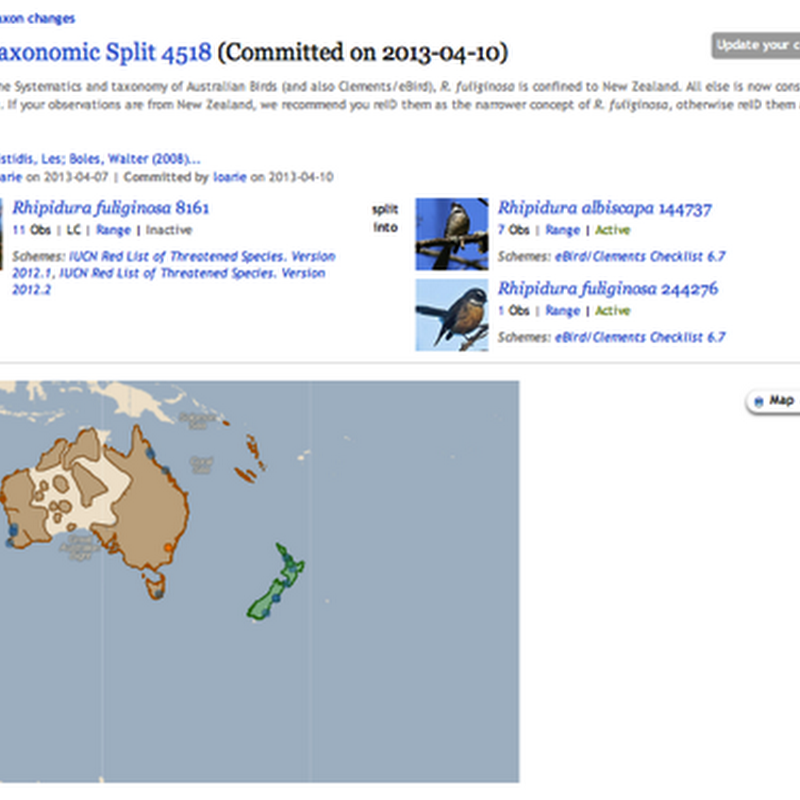
Donald Hobern drew my attention to nice the way iNaturalist displays taxonomic splits: In this example, observations identified as Rhipidura fuliginosa are being split into Rhipidura fuliginosa and Rhipidura albiscapa . This immediately reminds me of the idea which keeps circulating around, namely using version control tools to manage taxonomic classification.

Came across this paper recently: Despite QR Codes being uncool, there's something appealing about the idea of compressing a DNA barcode sequence into a small image.