
Given various discussions about identifiers, dark taxa, and DNA barcoding that have been swirling around the last few weeks, there's one notion that is starting to bug me more and more.

Given various discussions about identifiers, dark taxa, and DNA barcoding that have been swirling around the last few weeks, there's one notion that is starting to bug me more and more.
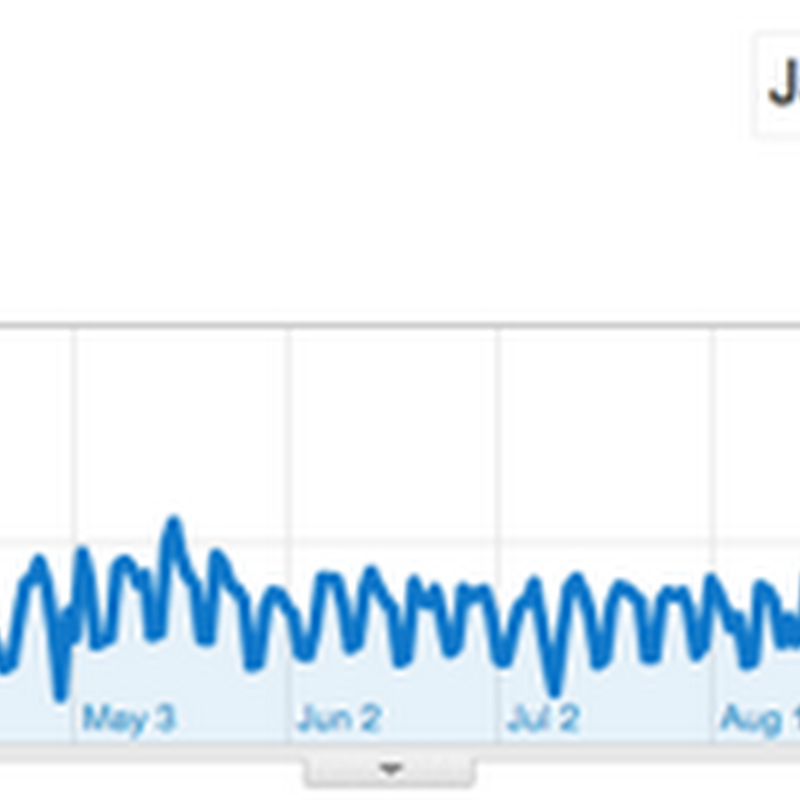
According to Google Analytics BioStor has experienced a big drop in traffic since the start of October: At one point I'm getting something like 4500 visits a week, now it's just over a thousand a week. I'm guessing this is due to Google's 'Panda' update. I suspect part of the problem is that in terms of text content BioStor is actually pretty thin.
The latest addition to my mapping of taxonomic names to the literature (http://iphylo.org/~rpage/itaxon/) is the ability for authors with Mendeley accounts to find the names they've published. This is an extension of the "I wrote that" tool I developed earlier. Let's say I want to show the names that a given author has published.
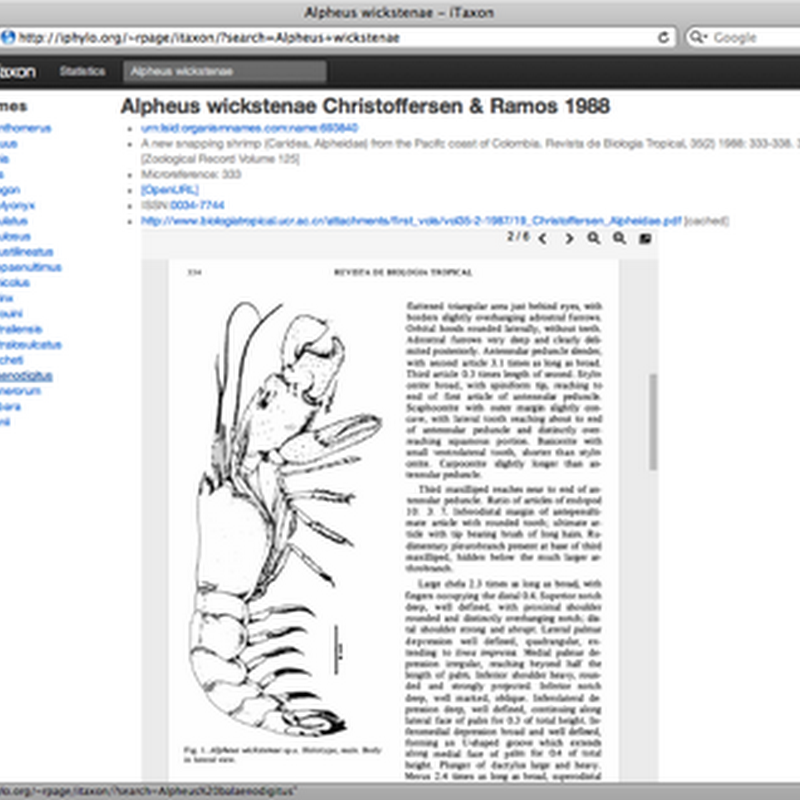
Following on from my earlier post Linking taxonomic names to literature: beyond digitised 5×3 index cards I've been slowly updating my latest toy: http://iphylo.org/~rpage/itaxon This site displays a database mapping over 200,000 animal names to the primary literature, using a mix of identifiers (DOIs, Handles, PubMed, URLs) as well as links to freely available PDFs where they are available.

David King et al.'s paper "Towards the bibliography of life" http://dx.doi.org/10.3897/zookeys.150.2167 has just appeared in a special issue of ZooKeys . I've written a number of posts on this topic, so I've a few comments. King et al. survey some of the issues, but don't really tackle the big issue of how we're going to build this.

Browsing EOL I stumbled upon the recently described fish Protoanguilla palau , shown below in an image by rairaiken2011: Two things struck me, the first is that the EOL page for this fish gives absolutely no clue as to where you would to find out more about this fish (apart from an unclickable link to the Wikipedia page http://en.wikipedia.org/wiki/Protoanguilla - seriously, a link that isn't clickable?), despite the fact this fish

Inspired by a comment on my post Visualising edit history of a Wikipedia page, the code I use to make history flow diagrams like the one below is now in GitHub at https://github.com/rdmpage/wikihistoryflow. There is also a live version at http://iphylo.org/~rpage/wikihistoryflow.
Quick note to self in case I (inevitably) forget later. If you are using Apache mod_rewrite to make nice, clean URLs, and are also supporting JSONP, you may run into the situation where you have code that wants to append "?callback=xxx" to your URL (e.g., a cross-domain AJAX call in jQuery). Imagine you have a nice clean URL /user/123, which actually corresponds to user.php?id=123.

Recently I've been thinking about the best ways to make article-level metadata from BioStor more widely available. For example, for someone visiting the BHL site there is no easy way to find articles, which are the basic unit for much of the scientific literature. How hard would it be to add articles to BHL?
One thing I'm increasingly conscious of is that I've a lot of demos and toy projects hanging around and the code for most of these isn't readily available. So, I plan to clean these up and put them in GitHub so others can explore the code, and reuse it if they see fit. First up is the code to create a HTML+Javascript clone of Nature's iPhone app, as described in an earlier post. There's a live version of the clone here here.