I stumbled across this paper (found on the GBIF Public Library):The first sentence of the abstract makes the paper sound a bit of a slog to read, but actually it's a great fun, full of pithy comments on the state of digital humanities. Almost all of this is highly relevant to mobilising natural history data.
iPhylo
I've been involved in a few Twitter exchanges about the upcoming pro-iBiosphere meeting regarding the "Open Biodiversity Knowledge Management System (OBKMS)", which is the topic of the meeting. Because for the life of me I can't find an explanation of what "Open Biodiversity Knowledge Management System" is, other than vague generalities and appeals to the magic pixie dust that is "Linked Open Data" and "RDF", I've been grumbling away on Twitter.
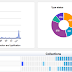
I'm adding more charts to the GBIF Chart tool, including some to explore the type status of specimens from the Solomon Islands.
Note to self on citation matching. Looking for this paper "Fishes of the Marshall and Marianas islands. Vol. I. Families from Asymmetrontidae through Siganidae" I Googled it, adding "bistro" as a search term to see if I'd already added it to BioStor.
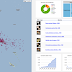
Following on from the previous post on visualising GBIF data, I've added some more interactivity.

Tim Roberston and the ream at GBIF are working on some nice visualisations of GBIF data, and have made an early release available for viewing: http://analytics.gbif-uat.org.

It is almost a year to the day that I released BioNames, a database of "taxa, texts, and trees". This project was my entry in EOL's Computable Data Challenge.
I had a long Twitter conversation with Terry Catapano (@catapanoth) today, and as can happen with a distracted stream of tweets, I think we were a little at cross purposes. This blog post is an attempt to unpack the debate.

As announced on phylobabble I've started to revisit visualising large phylogenies, building on some work I did a couple of years ago (my how time flies). This time, there is actual code (see https://github.com/rdmpage/deep-tree) as well as a live demo http://iphylo.org/~rpage/deep-tree/demo/. You can see the amphibian tree below at http://iphylo.org/~rpage/deep-tree/demo/show.php?id=5369171e32b7a:You can upload or paste a tree (for now in NEXUS
Some interesting threads in TAXACOM today (yes, really). The following article has appeared in Science :The authors argue that "The availability of adequate alternative methods of documentation, including high-resolution photography, audio recording, and nonlethal sampling, provide an opportunity to revisit and reconsider field collection practices and policies."This has brought a swift response from Kevin Winkler (Re)affirming the
The Biodiversity Heritage Library (BHL) has recently introduced a feature that I strongly dislike. The post describing this feature (Inspiring discovery through free access to biodiversity knowledge... states:What this means is that, whereas in the past a search in BHL would only turn up content actually in BHL, now that search may return results from other sources. What's not to like?