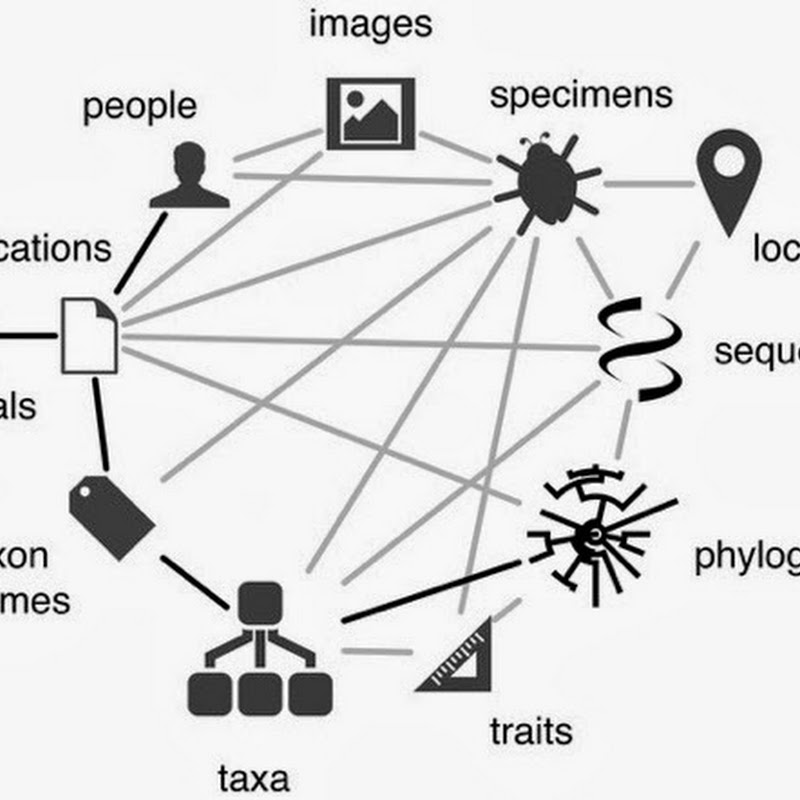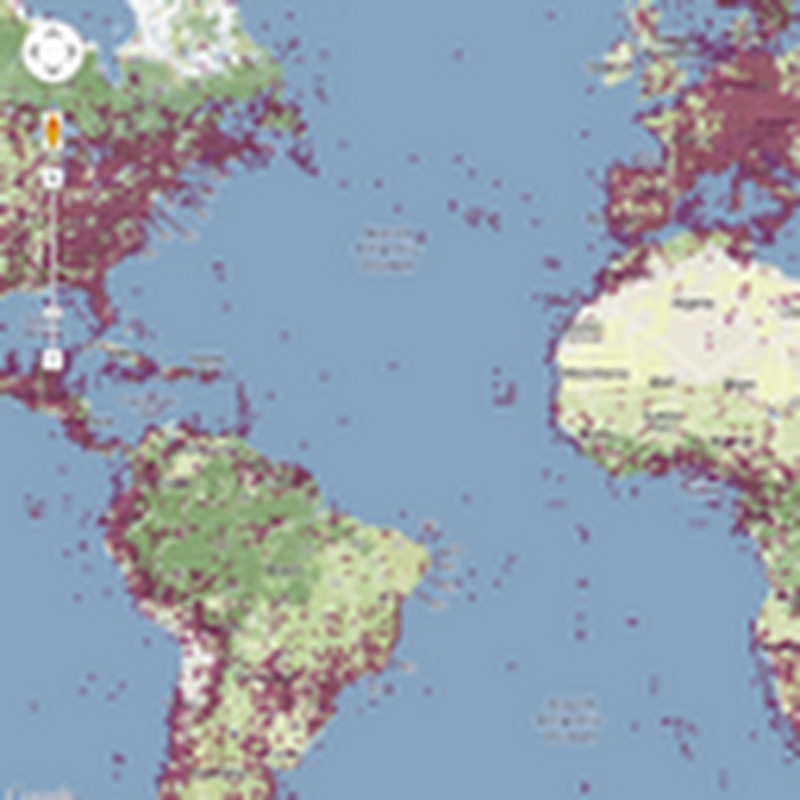
Following on from the previous post on putting GBIF data onto Google Maps, I'm now going to put DNA barcodes onto Google Maps. You can see the result at http://iphylo.org/~rpage/bold-map/, which displays around 1.2 million barcodes obtained from the International Barcode of Life Project (iBOL) releases.