I was working on a figure in Adobe Illustrator today. The ai file had 32 embedded TIFF files (we tend to embed images rather than linking them for portability reasons). I wanted to change all of the images, but to do this I needed to know where the originals were.
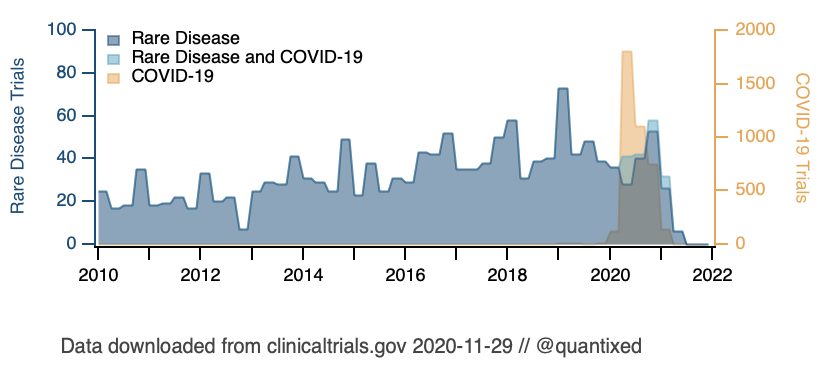
The database clinicaltrials.gov is a web resource of clinical trials around the world. It has a REST API that gives access to clinical trial data. There are some resources available to interact with this resource using R, such as rclinicaltrials and ClinicalTrialsAPI.

A short follow-up post. Previously, I looked at how to reproduce a Strava feature that compares performance over similar courses. With a few modifications to the code, I was able to analyse a much larger dataset of cycling performance on similar courses. Two courses with the highest number of tracks are shown below.
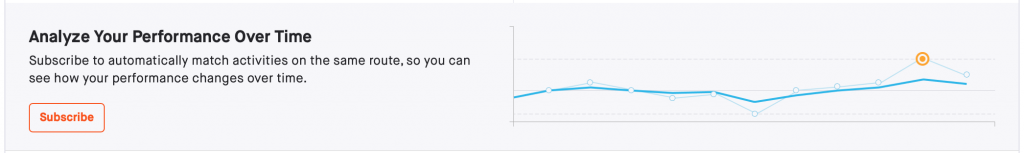
One of several features that Strava put behind a paywall was the ability to compare performance on similar courses. I miss this comparison tool and wondered how hard it would be to code my own. This post is a walkthrough of how I approached the problem. The code is available here.
We have a new preprint out. This is a short post to describe our findings and highlight some of the software I developed for analysing cell migration and cell shape data.
I searched several times in vain to solve this problem. After finding a solution, I thought i’d put it up here. The problem Formatting of units using siunitx in LaTeX does not match the typeface of the body text. We like to use the helvet package to get a close approximation to Helvetica in LaTeX.

Here is a fun post about using colour palettes in R. It starts with a computer game… After a few years of sporadically playing Super Mario World 2 – Yoshi’s Island on the Retropie, I made it to the final level.

During the pandemic, many virtual seminar programmes have popped up. One series, “Motors in Quarantine“, has been very successful. It’s organised by my colleagues Anne Straube, Alex Zwetsloot and Huong Vu. Anne wanted to know if attendees of the seminar series were a fair representation of the field.
The coronavirus crisis has meant that scientific meetings and seminars have moved online. This change has led to me wondering: why don’t scientists give talks the way that musicians do gigs? The idea is: after posting a preprint or publishing a paper, a scientist advertises that they will livestream a seminar to explain the work.

Joe Friel reposted an article earlier this year on Efficiency Factor in running. Efficiency Factor (EF) can be viewed in Training Peaks software and he describes how it is calculated. This post describes how I went about calculating EF in R using a single gpx file.

I’m a long-term fan of Weezer. Such was the brilliance of their first two albums that I have stuck with them through thick and thin. And dear me, there has been some very thin music. Nonetheless I own every album – thirteen of them.