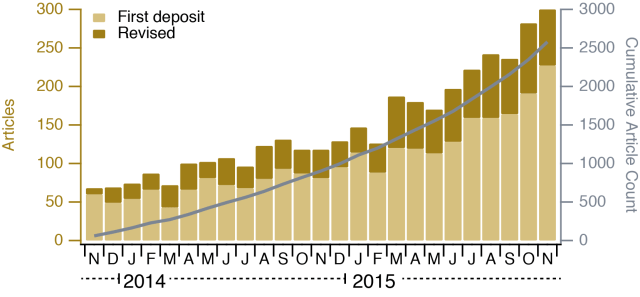
bioRxiv, the preprint server for biology, recently turned 2 years old. This seems a good point to take a look at how bioRxiv has developed over this time and to discuss any concerns sceptical people may have about using the service.

bioRxiv, the preprint server for biology, recently turned 2 years old. This seems a good point to take a look at how bioRxiv has developed over this time and to discuss any concerns sceptical people may have about using the service.

This post follows on from a previous post on citation distributions and the wrongness of Impact Factor.

{.alignright .size-medium .wp-image-551 loading=“lazy” decoding=“async” attachment-id=“551” permalink=“https://quantixed.org/jcbdiet/” orig-file=“https://i0.wp.com/quantixed.org/wp-content/uploads/2015/07/jcbdiet.jpg?fit=603%2C2586&ssl=1” orig-size=“603,2586” comments-opened=“1”

This is a long post about Journal Impact Factors. Thanks to Stephen Curry for encouraging me to post this.

I saw this great tweet (fairly) recently: I thought this was such a great explanation of when to submit your paper. It reminded me of a diagram that I sketched out when talking to a student in my lab about a paper we were writing. I was trying to explain why we don’t exaggerate our findings. And conversely why we don’t undersell our results either. I replotted it below:
Our most recent manuscript was almost ready for submission. We were planning to send it to an open access journal. It was then that I had the thought: how many papers in the reference list are freely available? It somehow didn’t make much sense to point readers towards papers that they might not be able to access. So, I wondered if there was a quick way to determine how papers in my reference list were open access.

Following on from the last post about publication lag times at cell biology journals, I went ahead and crunched the numbers for all journals in PubMed for one year (2013). Before we dive into the numbers, a couple of points about this kind of information. Some journals “reset the clock” on the received date with manuscripts that are resubmitted. This makes comparisons difficult.

My interest in publication lag times continues. Previous posts have looked at how long it takes my lab to publish our work, how often trainees publish and I also looked at very long lag times at Oncogene. I recently read a blog post on automated calculation of publication lag times for Bioinformatics journals. I thought it would be great to do this for Cell Biology journals too.

I thought I’d share this piece of analysis looking at productivity of people in the lab. Here, productivity means publishing papers. This is unfortunate since some people in my lab have made some great contributions to other peoples’ projects or have generally got something going, but these haven’t necessarily transferred into print. Also, the projects people have been involved in have varied in toughness.