A new version of rentrez, our package for the NCBI’s EUtils API, is makingit’s way around the CRAN mirrors. This release represents a substantialimprovement to rentrez, including a new vignettethat documents the whole package. This posts describes some of the new things in rentrez, and gives us a chanceto thank some of the people that have contributed to this package’s development.
We’re happy to announce the launch of a CRAN-style repository for rOpenSci at http://packages.ropensci.org This repository contains the latest nightly builds from the master branch of all rOpenSci packages currently on GitHub.
Despite the hype around “big data”, a more immediate problem facing many scientific analyses is that large-scale databases must be assembled from a collection of small independent and heterogeneous fragments – the outputs of many and isolated scientific studies conducted around the globe. Collecting and compiling these fragments is challenging at both political and technical levels.
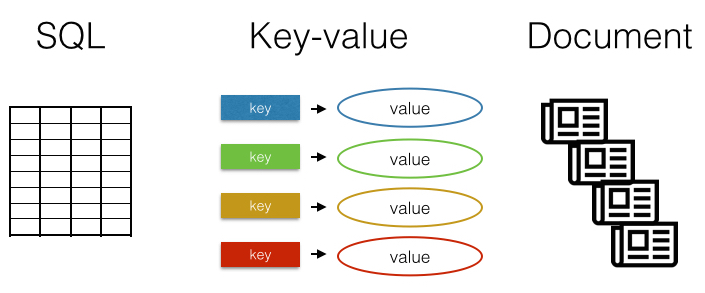
There are many different databases. The most familiar are row-column SQL databases like MySQL, SQLite, or PostgreSQL. Another type of database is the key-value store, which as a concept is very simple: you save a value specified by a key, and you can retrieve a value by its key. One more type is the document database, which instead of storing rows and columns, stores blobs of text or even binary files.
There are two things that make R such a wonderful programming environment - the vast number of packages to access, process and interpretdata, and the enthusiastic individuals and subcommunities (of which rOpenSci is a great example). One, of course, flows from the other:R programmers write R packages to provide language users with more features, which makes everyone’s jobs easier and (hopefully!)attracts more users and more contributions.
rOpenSci specializes in creating R libraries for accessing data resources on the web from R. Most times you request data from the web in R with our packages, you should have no problem. However, you evenutally will run into problems. In addition, there are advanced things you can do modifying requests to web resources that fall in the advanced stuff category.
Key to the success of rOpenSci is our community and we want to hear more regularly from our members, and foster new interactions among the group. In addition, community calls are a way for us to give important updates, and get feedback on them. We tentatively plan on doing community calls once per month. The format of rOpenSci community calls could be of various types.

Why open data growth At rOpenSci we try to make it easier for people to use open data and contribute open data to the community. The question often arises: How much open data do we have? Another angle on this topic is: How much is open data growing? We provide access to dozens of data respositories through our various packages.
So what is Docker? Docker is a relatively new opensource applicationand service, which is seeing interest across a number of areas. Ituses recent Linux kernel features (containers, namespaces) to shieldprocesses.
I’m very pleased to announce that rOpenSci has signed a comprehensive fiscal sponsorship agreement with the NumFocus foundation, a 501(c)3 nonprofit that supports R&D for open source scientific software projects.
The week after labor day, we had the pleasure of attending the NCEAS open science codefest event in Santa Barbara. It was great to meet folks like the new arrivals at the expanding Mozilla Science Lab, Bill Mills and Abby Cabunoc (Bill even already has a great post up about the codefest), and see old friends from NCEAS and DataONE, among many more. This 2.5 day event ran smoothly thanks to the leadership of Matt Jones.