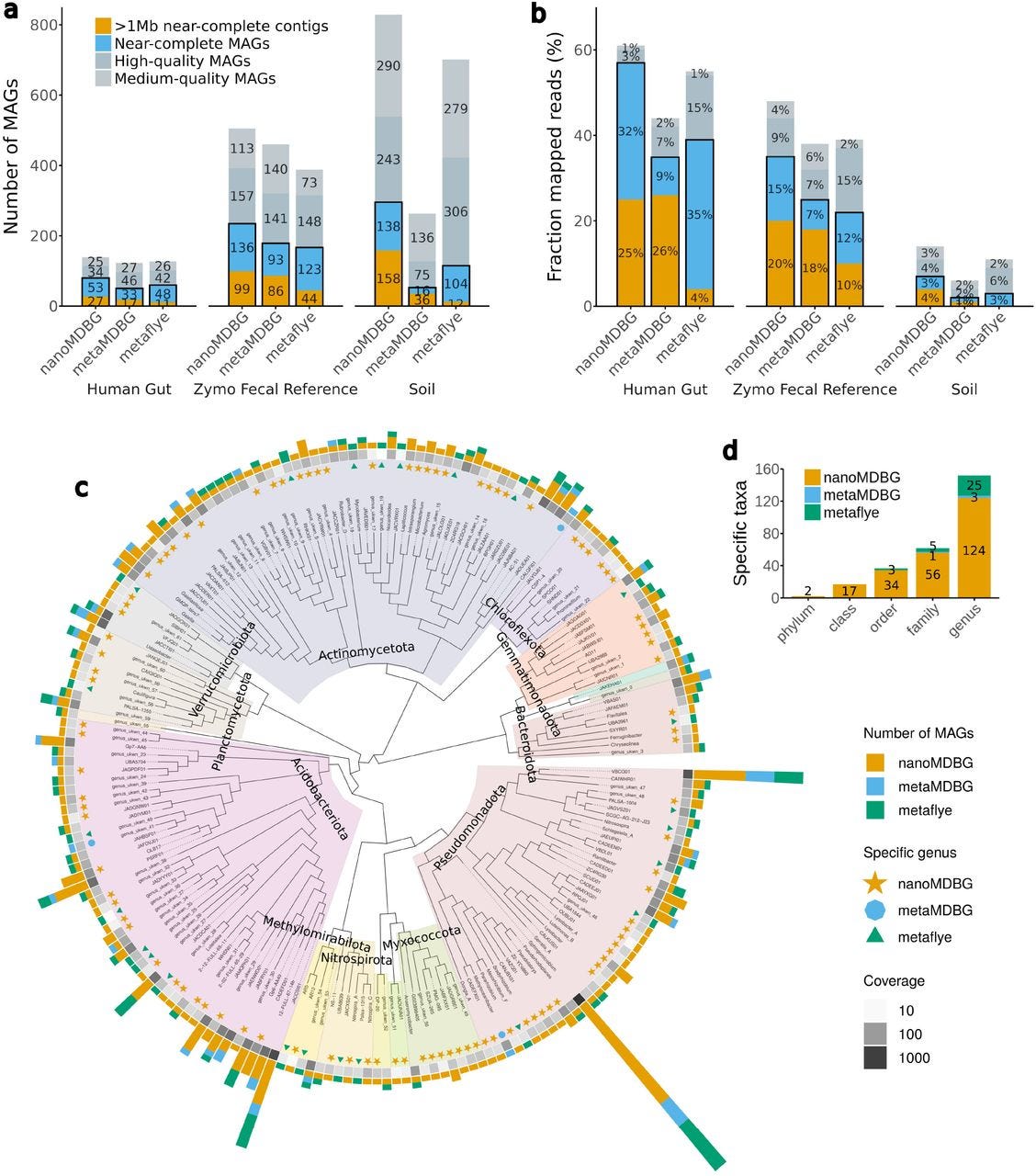
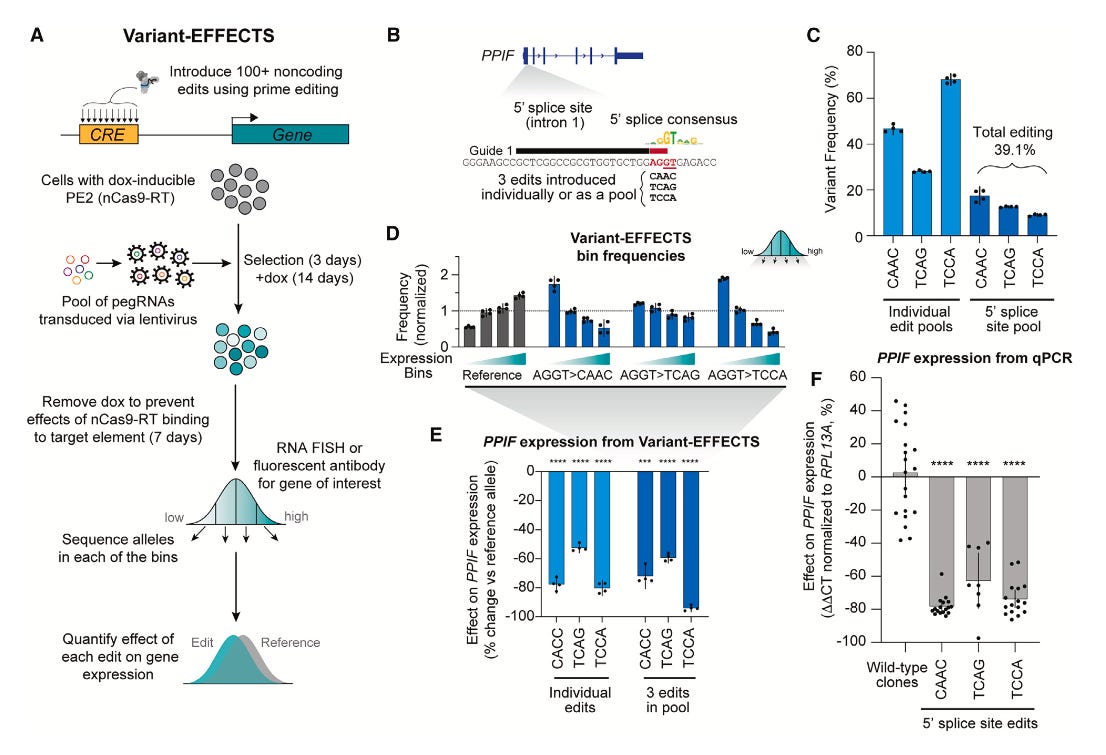
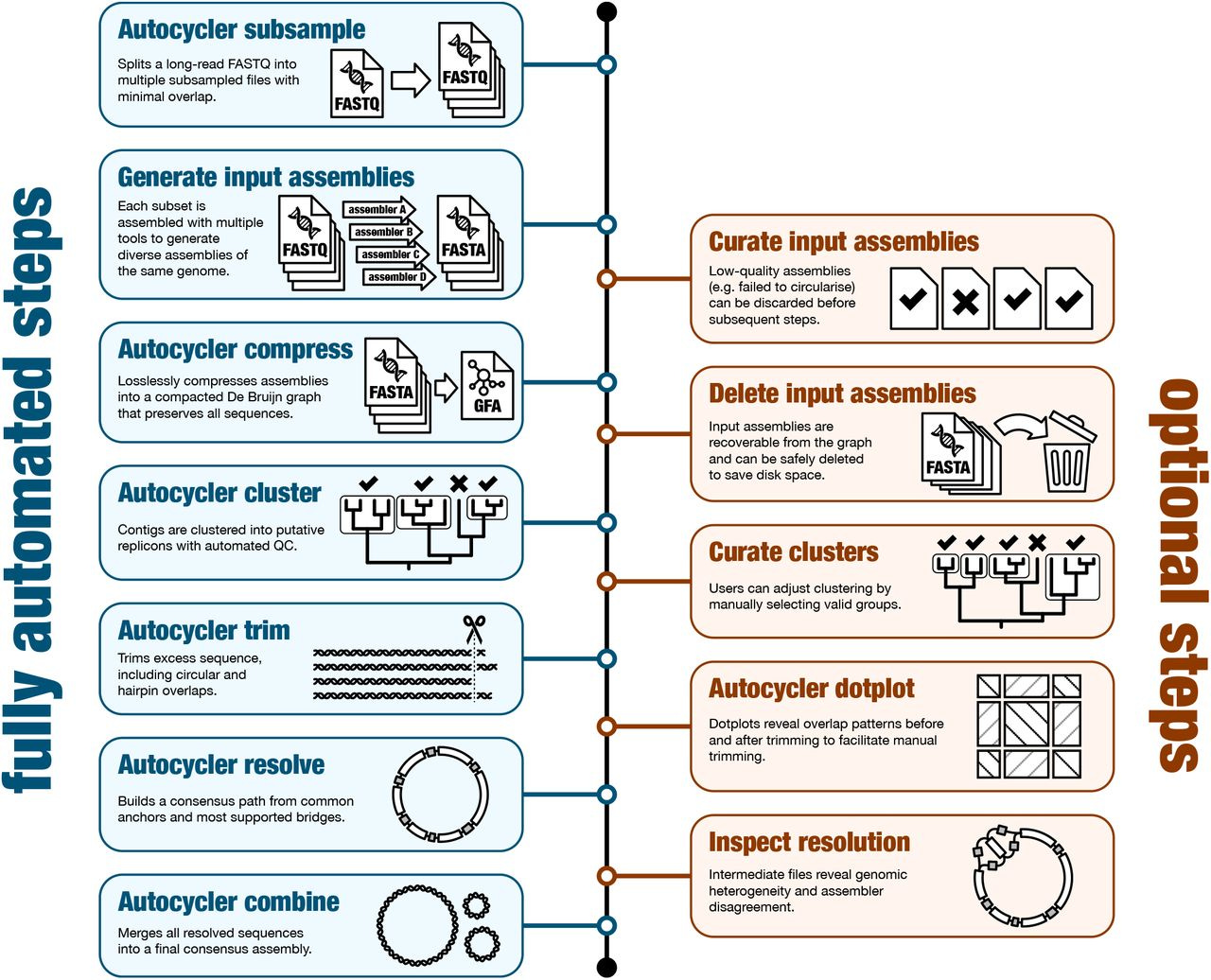
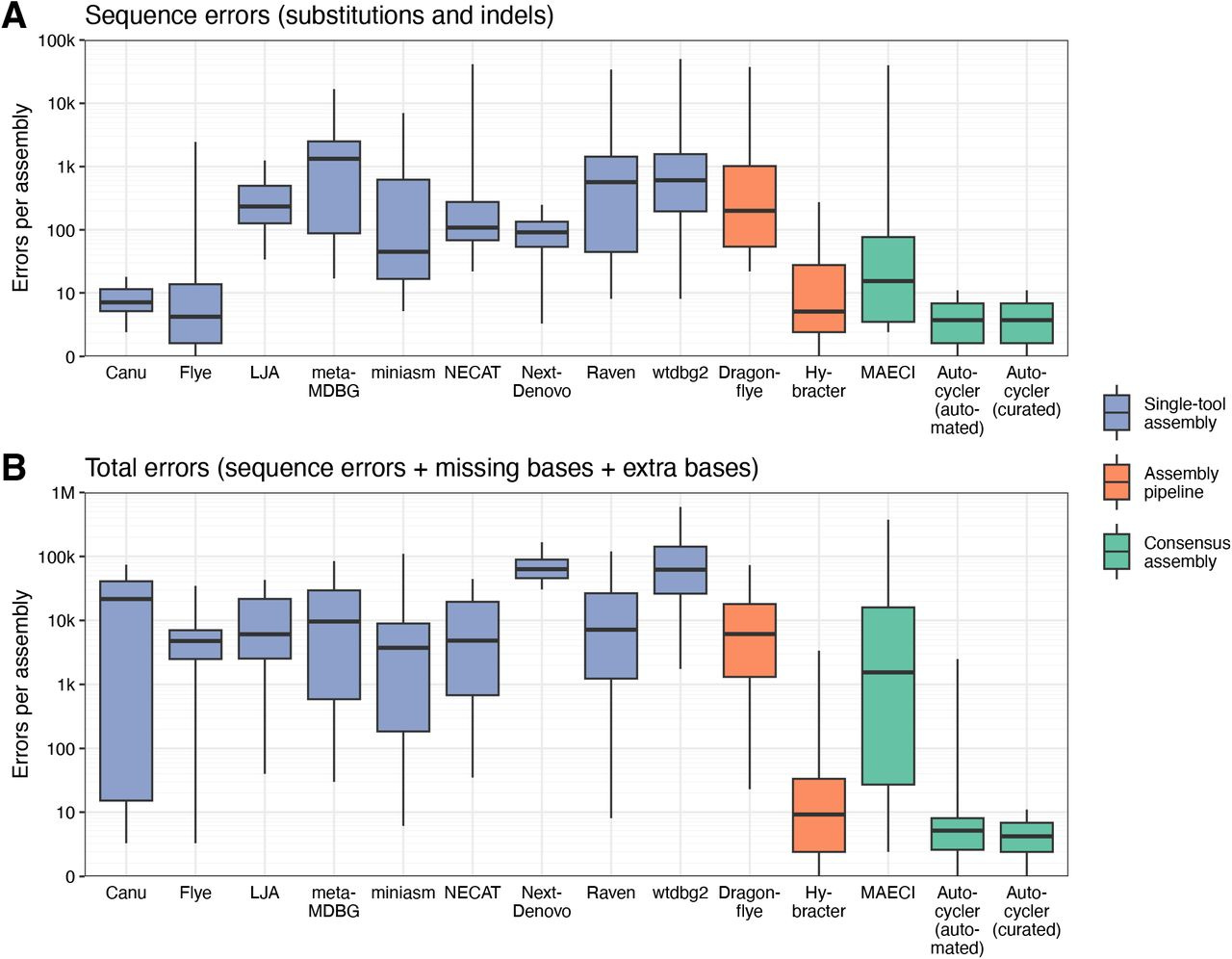
I recently wrote a piece about leaving academia for biotech. I left academia for industry in 2019. I spent four years at a consulting firm before joining Colossal Biosciences. This week I’m returning to the University of Virginia School of Data Science as a tenured associate professor and dean of research. The transition from academia to industry can be tricky, but it’s also increasingly common.