TL;DR: Codeium offers a free Copilot-like experience in Positron. You can install it from the Open VSX registry directly within the extensions pane in Positron.

TL;DR: Codeium offers a free Copilot-like experience in Positron. You can install it from the Open VSX registry directly within the extensions pane in Positron.

Last month I published a paper and an R package for summarizing preprints from bioRxiv using a local LLM. I wrote about it here: Llama 3.2 was just released today (announcement). The biggest news is the addition of a multimodal vision model, but I was intrigued by the reasonably good performance of the tiny 3B text model. I used this as an excuse to update the biorecap R package.
Llama 3.1 405B is the first open-source LLM on par with frontier models GPT-4o and Claude 3.5 Sonnet. I’ve been running the 70B model locally for a while now using Ollama + Open WebUI, but you’re not going to run the 405B model on your MacBook.
VP (Pete) Nagraj is a long time friend, colleague, and collaborator, and is the author of this post. Pete and I have co-authored over a dozen publications, and have taught several graduate courses in data science together. Pete leads the health security analytics / infectious disease modeling and forecasting group at Signature Science, where we started this work together last year.

TL;DR I wrote an R package that summarizes recent bioRxiv preprints using a locally running LLM via Ollama+ollamar, and produces a summary HTML report from a parameterized RMarkdown template.
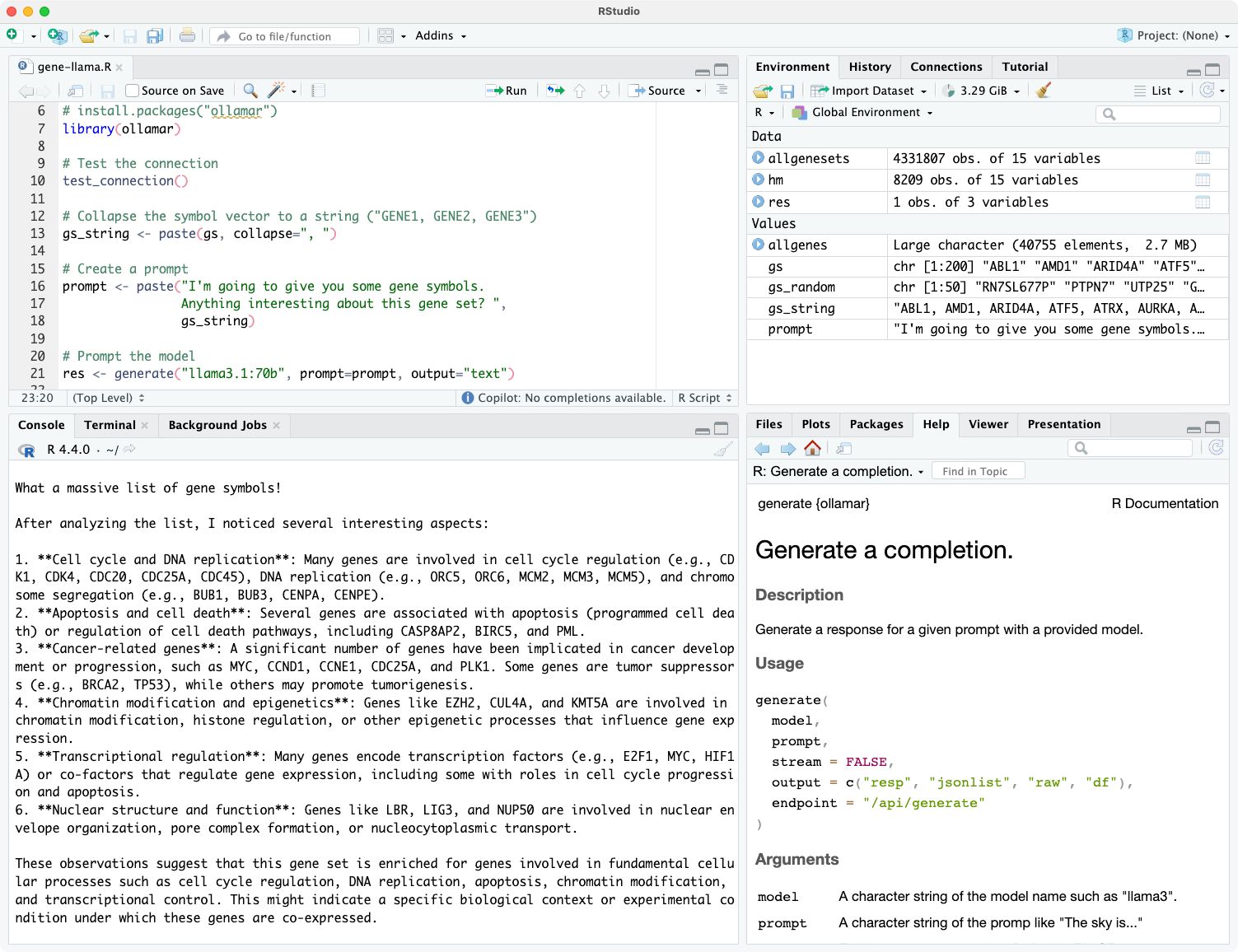
I’ve been using the llama3.1:70b model just released by Meta using Ollama running on my MacBook Pro. Ollama makes it easy to talk to a locally running LLM in the terminal (ollama run llama3.1:70b) or via a familiar GUI with the open-webui Docker container. Here I’ll demonstrate using the ollamar package on CRAN to talk to an LLM running locally on my Mac.
This post is about the R package development experience with Positron, the new IDE from Posit based on VS Code. This is not a tutorial on R package development in general — there are great resources for that elsewhere. Read on.Subscribe to Paired Ends to get future posts like this delivered to your e-mail. RStudio, VS Code, and Positron Back in 2011 I wrote a blog post about a relatively new IDE for R called RStudio.