
TL;DR I wrote an R package that summarizes recent bioRxiv preprints using a locally running LLM via Ollama+ollamar, and produces a summary HTML report from a parameterized RMarkdown template.

TL;DR I wrote an R package that summarizes recent bioRxiv preprints using a locally running LLM via Ollama+ollamar, and produces a summary HTML report from a parameterized RMarkdown template.
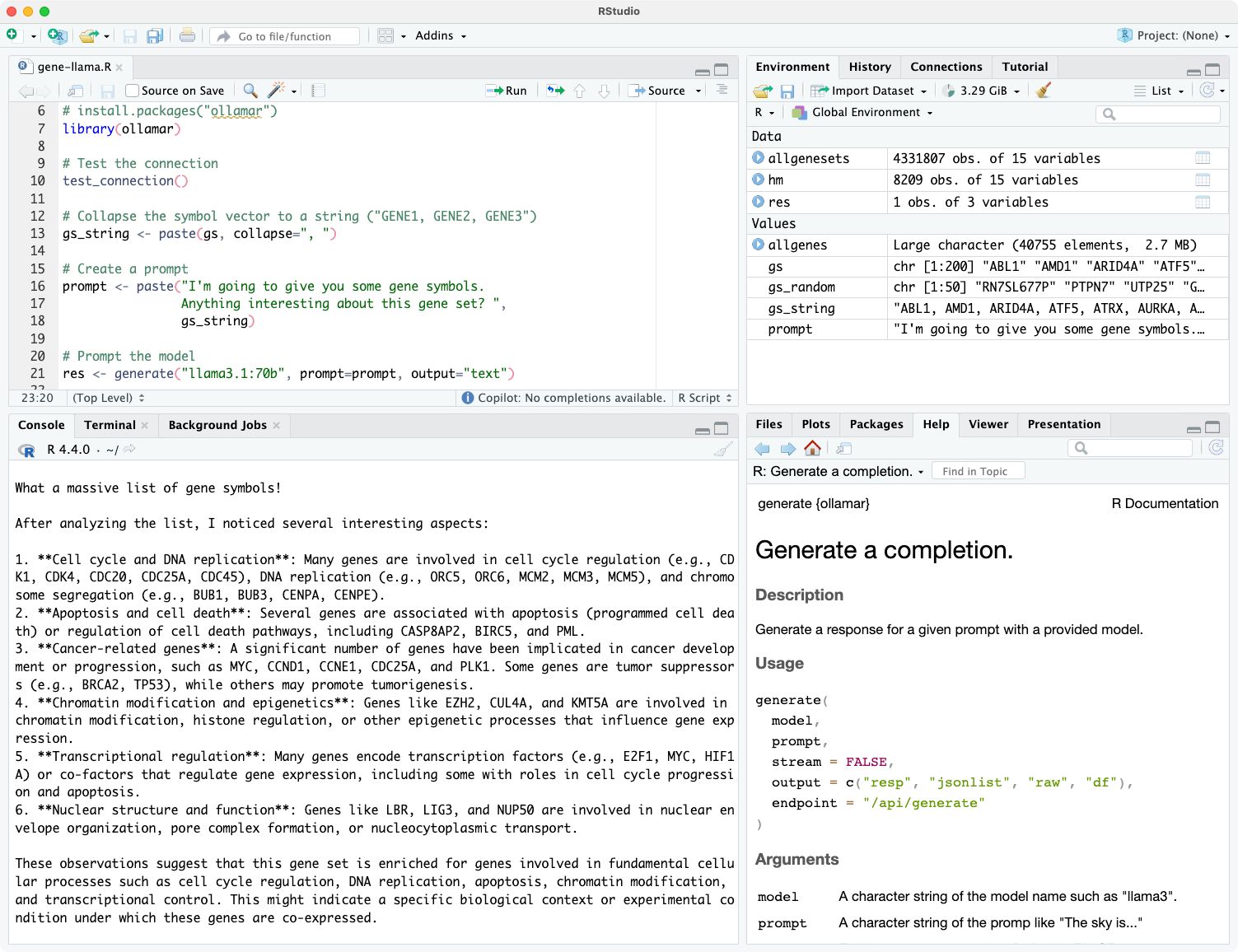
I’ve been using the llama3.1:70b model just released by Meta using Ollama running on my MacBook Pro. Ollama makes it easy to talk to a locally running LLM in the terminal (ollama run llama3.1:70b) or via a familiar GUI with the open-webui Docker container. Here I’ll demonstrate using the ollamar package on CRAN to talk to an LLM running locally on my Mac.
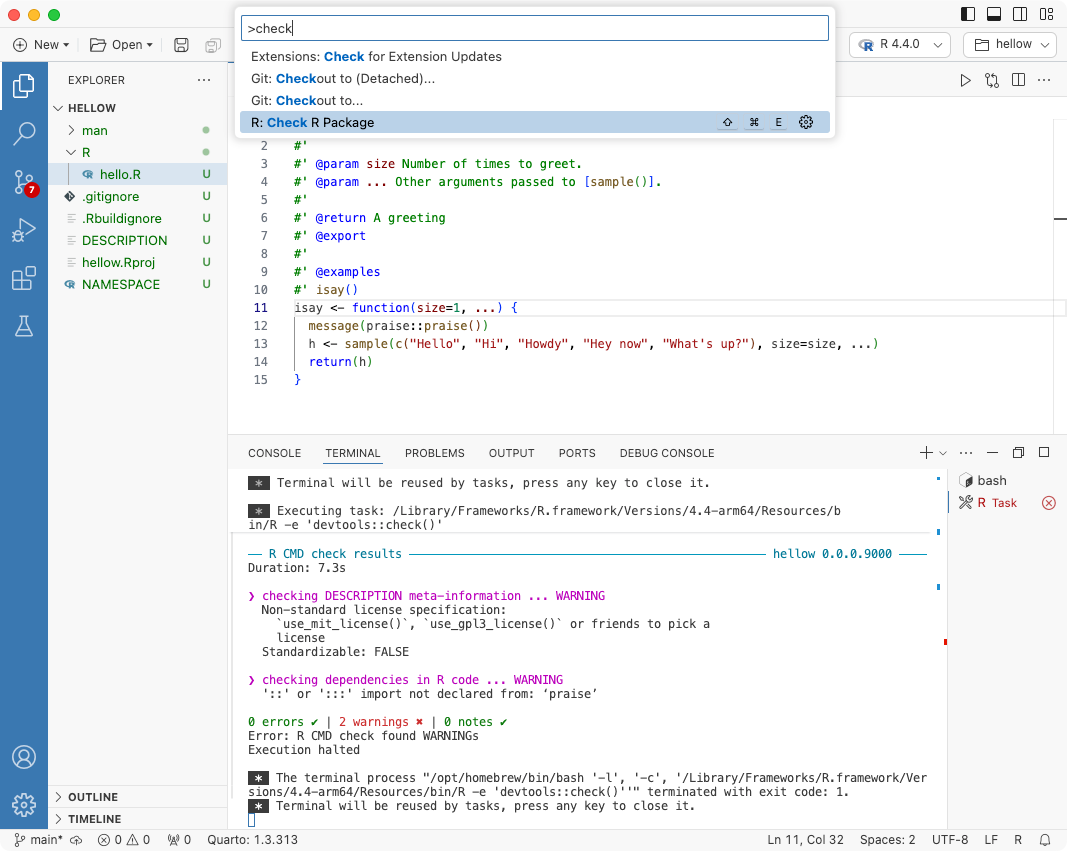
This post is about the R package development experience with Positron, the new IDE from Posit based on VS Code. This is not a tutorial on R package development in general — there are great resources for that elsewhere. Read on.Subscribe to Paired Ends to get future posts like this delivered to your e-mail. RStudio, VS Code, and Positron Back in 2011 I wrote a blog post about a relatively new IDE for R called RStudio.