
AI creates an education challenge, not just a job crisis. We haven't built systems to help people to continue learning and connect them to new opportunities.

AI creates an education challenge, not just a job crisis. We haven't built systems to help people to continue learning and connect them to new opportunities.

If an AI produces something useful it's because of your own skill in model choice, prompting, and steering. If not, it's because the model is a useless lying machine that can't follow directions.
R updates (R Data Scientist, R Weekly), AI accelerating wet lab bio research, Docker hardened images, LLMs in review, machinal bypass, science funding, new papers.

The "Machinal Bypass," when AI becomes a shortcut around the work that makes us human, and why some tasks should feel a little hard.
Writing better code without AI, AI in peer review, local LLMs, Biothreat Benchmark Generation, R updates (R Data Scientist, RWeekly, R Works), red-teaming an AI vending machine, new papers

Biotech policy: Recent policy discussions echo NSCEB recommendations across investment, defense, and data.
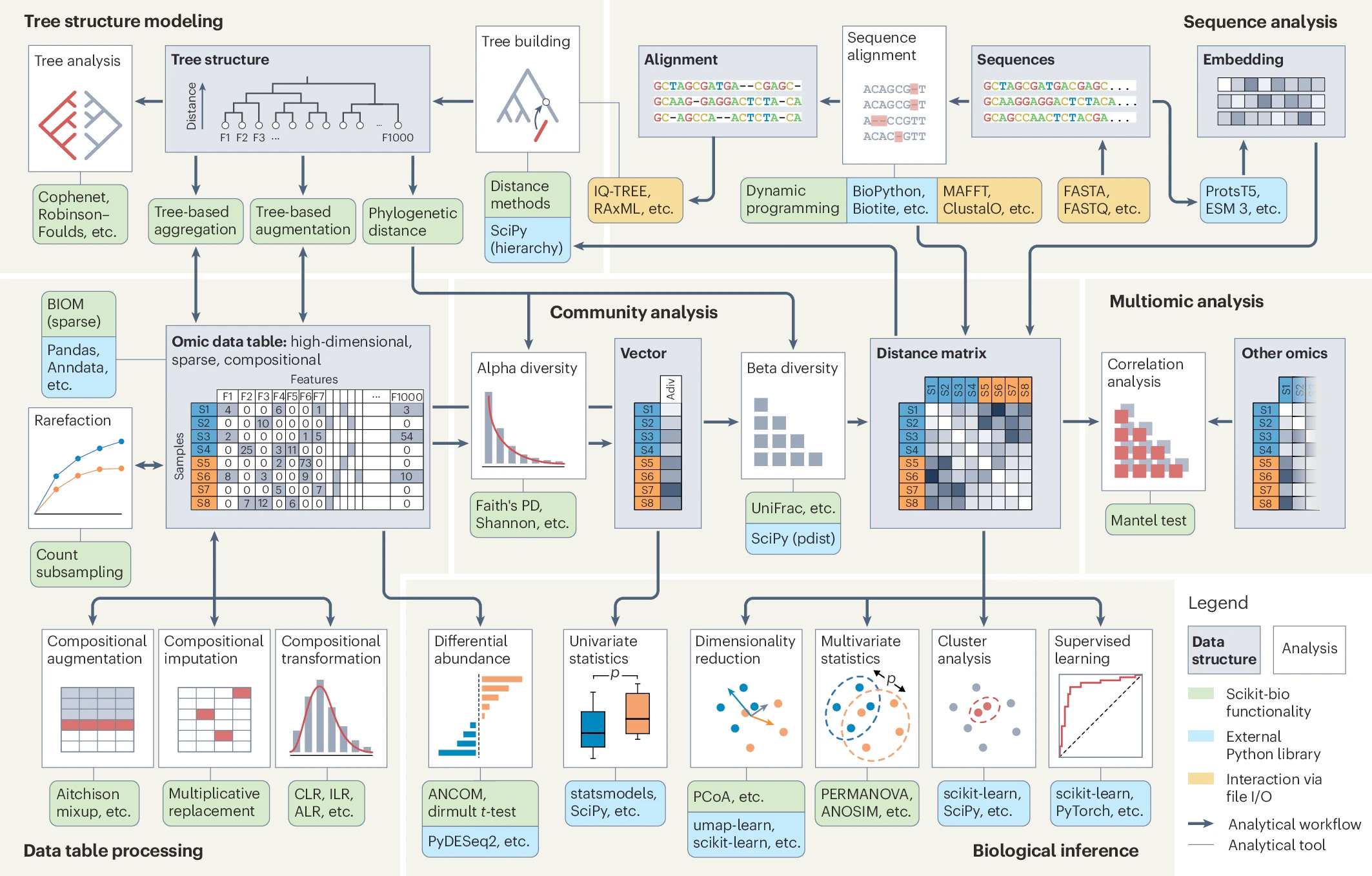
The "missing middle" for omics data analysis in Python
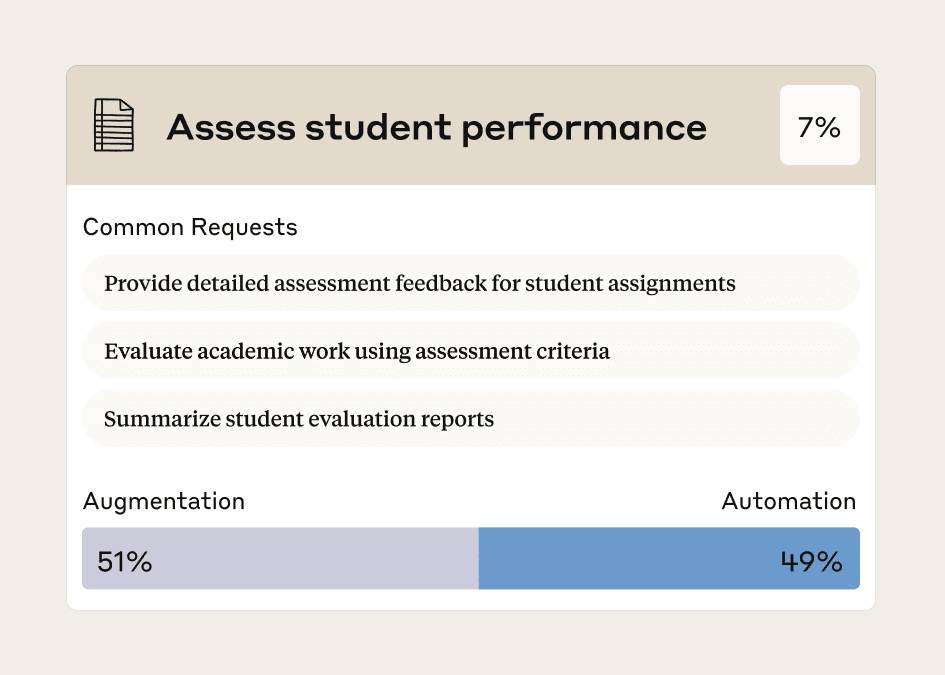
The Anthropic Education Report showed 50% of Claude conversations about grading delegated assessment to the AI. How does one actually design AI-resistant assignments?

"I did not sufficiently counterbalance that with skepticism and verification at the moment it mattered... I did not enforce the standard you expect: only claim access to evidence I can actually see"
GPT-5.2, DARPA Generative Optogenetics, don't use local models for coding agents, red teaming, sandbagging, AI in science &
Five principles for the future of computing: private, dedicated, plural, adaptable, and prosocial.