
I put a recent code snippet put up on the IgorExchange. It’s a simple procedure for averaging a set of 1D waves and putting the results in a new wave.

I put a recent code snippet put up on the IgorExchange. It’s a simple procedure for averaging a set of 1D waves and putting the results in a new wave.
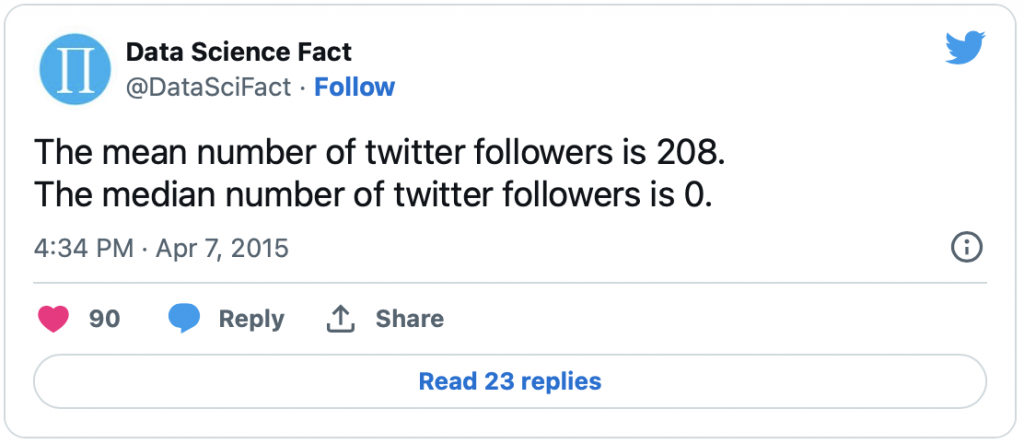
This is a long post about Journal Impact Factors. Thanks to Stephen Curry for encouraging me to post this. tl;dr — I really liked this recent tweet from Stat Fact It’s a great illustration of why reporting means for skewed distributions is a bad idea.
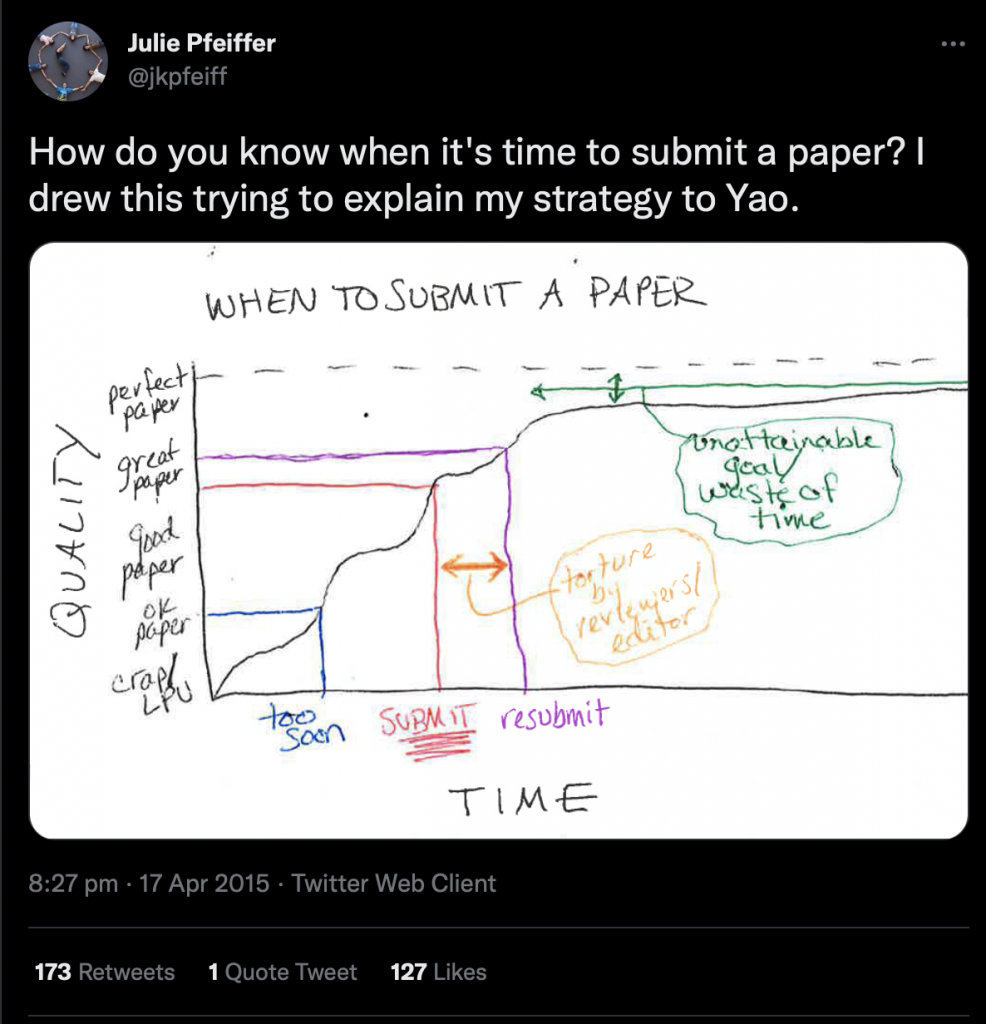
I saw this great tweet (fairly) recently: I thought this was such a great explanation of when to submit your paper. It reminded me of a diagram that I sketched out when talking to a student in my lab about a paper we were writing. I was trying to explain why we don’t exaggerate our findings.
We were asked to write a Preview piece for Developmental Cell. Two interesting papers which deal with the insertion of amphipathic helices in membranes to influence membrane curvature during endocytosis were scheduled for publication and the journal wanted some “front matter” to promote them.
Our most recent manuscript was almost ready for submission. We were planning to send it to an open access journal. It was then that I had the thought: how many papers in the reference list are freely available? It somehow didn’t make much sense to point readers towards papers that they might not be able to access.
Following on from the last post about publication lag times at cell biology journals, I went ahead and crunched the numbers for all journals in PubMed for one year (2013). Before we dive into the numbers, a couple of points about this kind of information.
My interest in publication lag times continues. Previous posts have looked at how long it takes my lab to publish our work, how often trainees publish and I also looked at very long lag times at Oncogene. I recently read a blog post on automated calculation of publication lag times for Bioinformatics journals.
I thought I’d share this piece of analysis looking at productivity of people in the lab. Here, productivity means publishing papers.
I thought I’d compile a list of songs related to biomedical science. These were all found in my iTunes library. I’ve missed off multiple entries for the same kind of thing, as indicated.

In this post I’ll describe a computational method for splitting two sides of a cell biological structure. It’s a simple method that relies on principal component analysis, otherwise known as PCA.
We have a new paper out! You can access it here. The work was mainly done by Cristina Gutiérrez Caballero, a post-doc in the lab. We had some help from Selena Burgess and Richard Bayliss at the University of Leicester, with whom we have an ongoing collaboration.