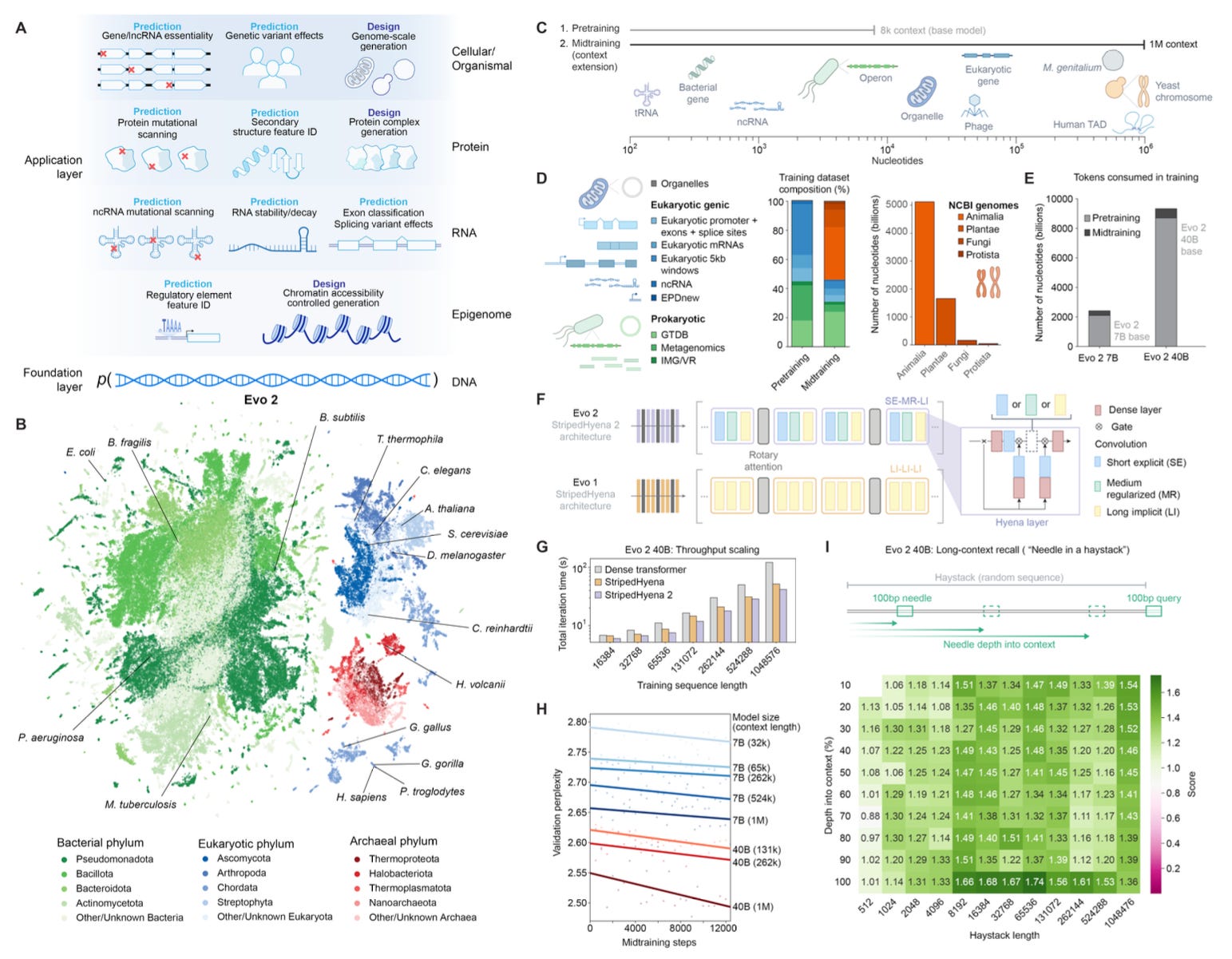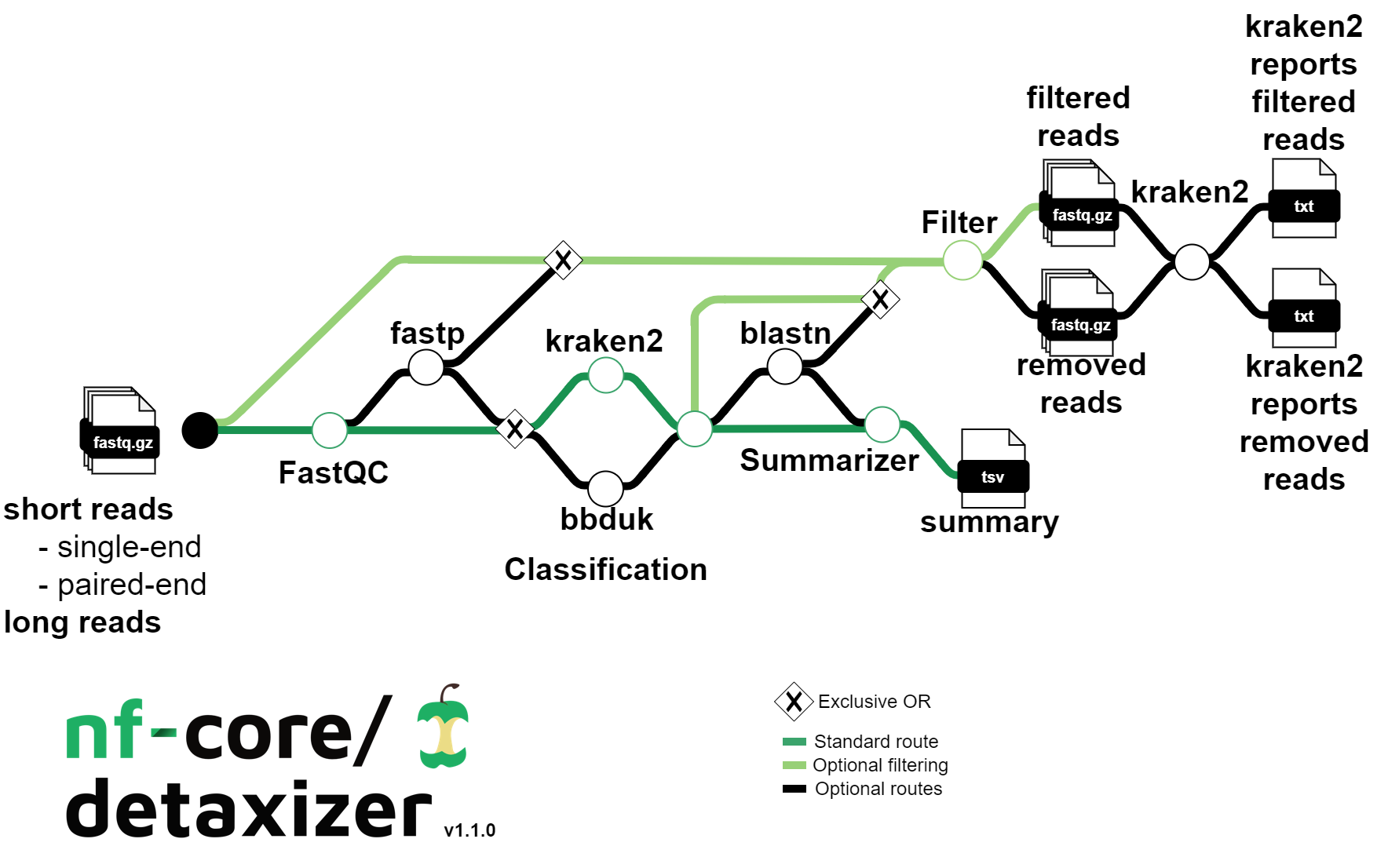
This week’s recap highlights polars-bio for fast and scalable and out-of-core operations on large genomic interval datasets, combining DNA and protein alignments to improve genome annotation with LiftOn, feature selection methods for scRNA-seq, STRkit for read-level genotyping of short tandem repeats using long reads and single-nucleotide variation, and nf-core/detaxizer for decontamination of human sequences in metagenomics data.