The Biodiversity Heritage Library (BHL) has recently introduced a feature that I strongly dislike. The post describing this feature (Inspiring discovery through free access to biodiversity knowledge... states:What this means is that, whereas in the past a search in BHL would only turn up content actually in BHL, now that search may return results from other sources. What's not to like?
iPhylo

Following on from earlier posts on annotating biodiversity data (Rethinking annotating biodiversity data and More on annotating biodiversity data: beyond sticky notes and wikis) I've started playing with user interfaces for editing data.
An undergraduate student (Aime Rankin) doing a project with me on citation and impact of museum collections came across a paper I hadn't seen before:Unfortunately the paper is behind a paywall, but here's the abstract (you can also get a PDF here):It's well worth a read. It argues that sequence databases such as Genbank are essentially the equivalent of the great natural history museums of the 19th Century. There are several ironies here.
Following on from the previous post Rethinking annotating biodiversity data, here are some more thoughts on annotating biodiversity data. Annotations as sticky notes I get the sense that most people think of annotations as "sticky notes" that someone puts on data. In other words, the data is owned by somebody, and anyone who isn't the owner gets to make comments, which the owner is free to use or ignore as they see fit.
TL;DR By using bookmarklets and a central annotation store, we can build a system to annotate any biodiversity database, and display those annotations on those databases. A couple of weeks ago I was at GBIF meeting in Copenhagen, and there was a discussion about adding a new feature to the GBIF portal. The conversation went something like this: Resources are limited, and adding new features to a project can be difficult.

Today I managed to publish some data from a GitHub repository directly to GBIF. Within a few minutes (and with Tim Robertson on hand via Skype to debug a few glitches) the data was automatically indexed by GBIF and its maps updated. You can see the data I uploaded here.The data I uploaded came from this paper:This is the data I used to build the geophylogeny for Banza using Google Earth.
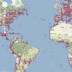
Following on from the previous post on putting GBIF data onto Google Maps, I'm now going to put DNA barcodes onto Google Maps. You can see the result at http://iphylo.org/~rpage/bold-map/, which displays around 1.2 million barcodes obtained from the International Barcode of Life Project (iBOL) releases.

As part of a project exploring GBIF data I've been playing with displaying GBIF data on Google Maps.

A quick note to myself to document a problem with the GBIF classification of liverworts (I've created issue POR-1879 for this).While building a new tool to browse GBIF data I ran into a problem that the taxon "Jungermanniales" popped up in two different places in the GBIF classification, which broke a graphical display widget I was using.If you search GBIF for Jungermanniales you get two results, both listed as "accepted":Based on Wikipedia

There is a great post by Jeni Tennison on the Open Data Institute blog entitled Five Stages of Data Grief.

I gave a remote presentation at a proiBioSphere workshop this morning. The slides are below (to try and make it a bit more engaging than a desk of Powerpoints I played around with Prezi).There is a version on Vimeo that has audio as well.I sketched out the biodiversity "knowledge graph", then talked about how mark-up relates to this, finishing with a few questions.