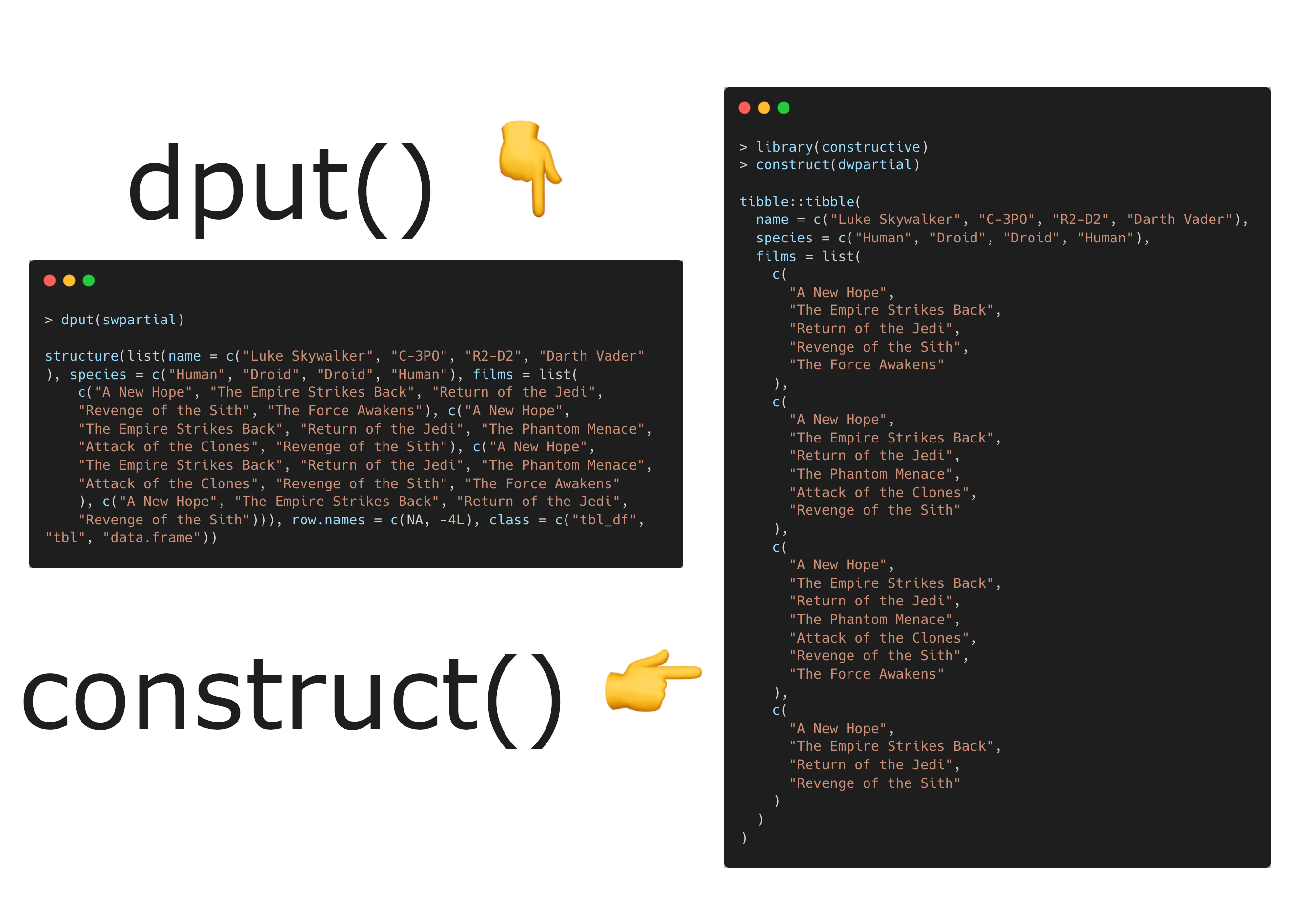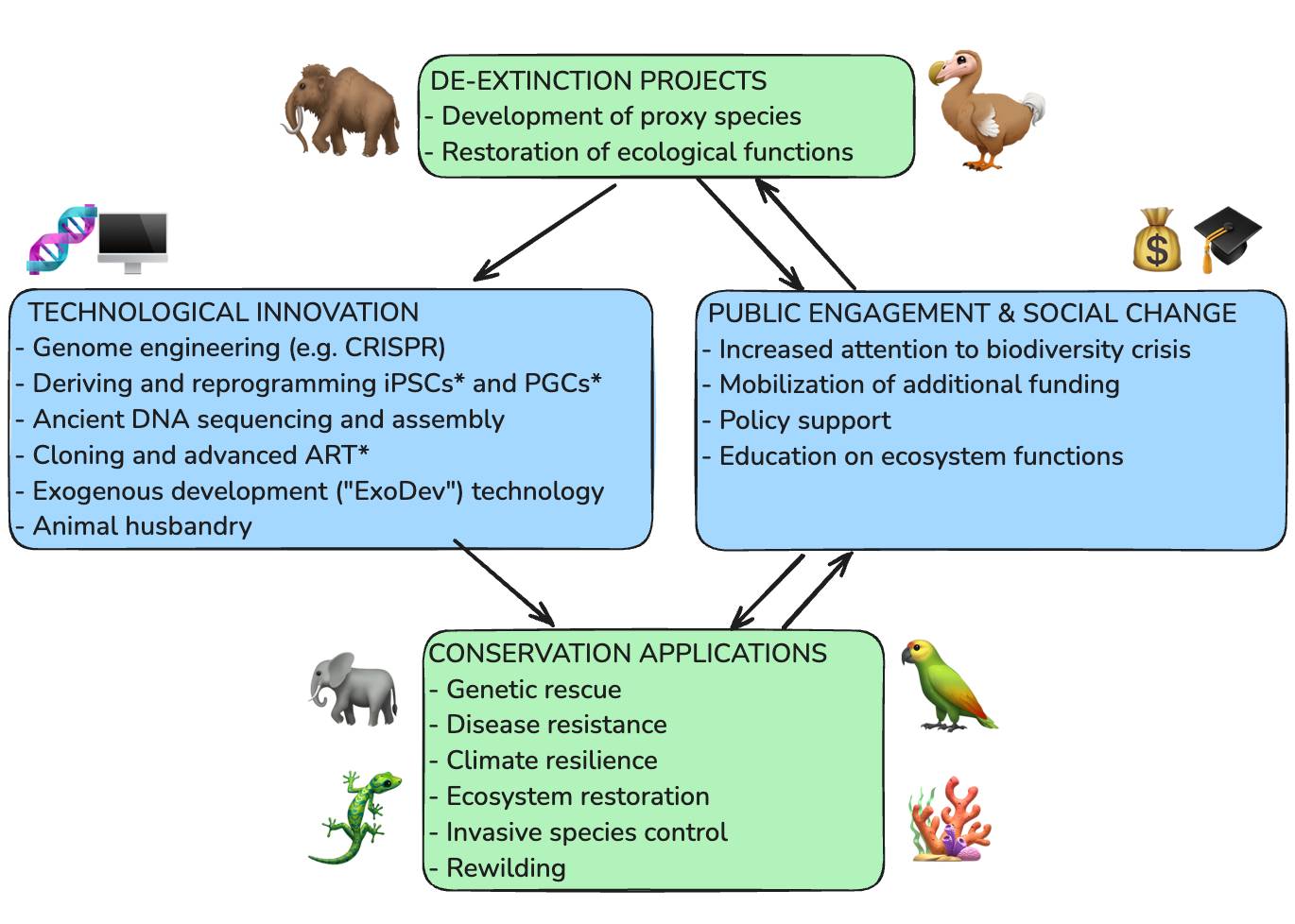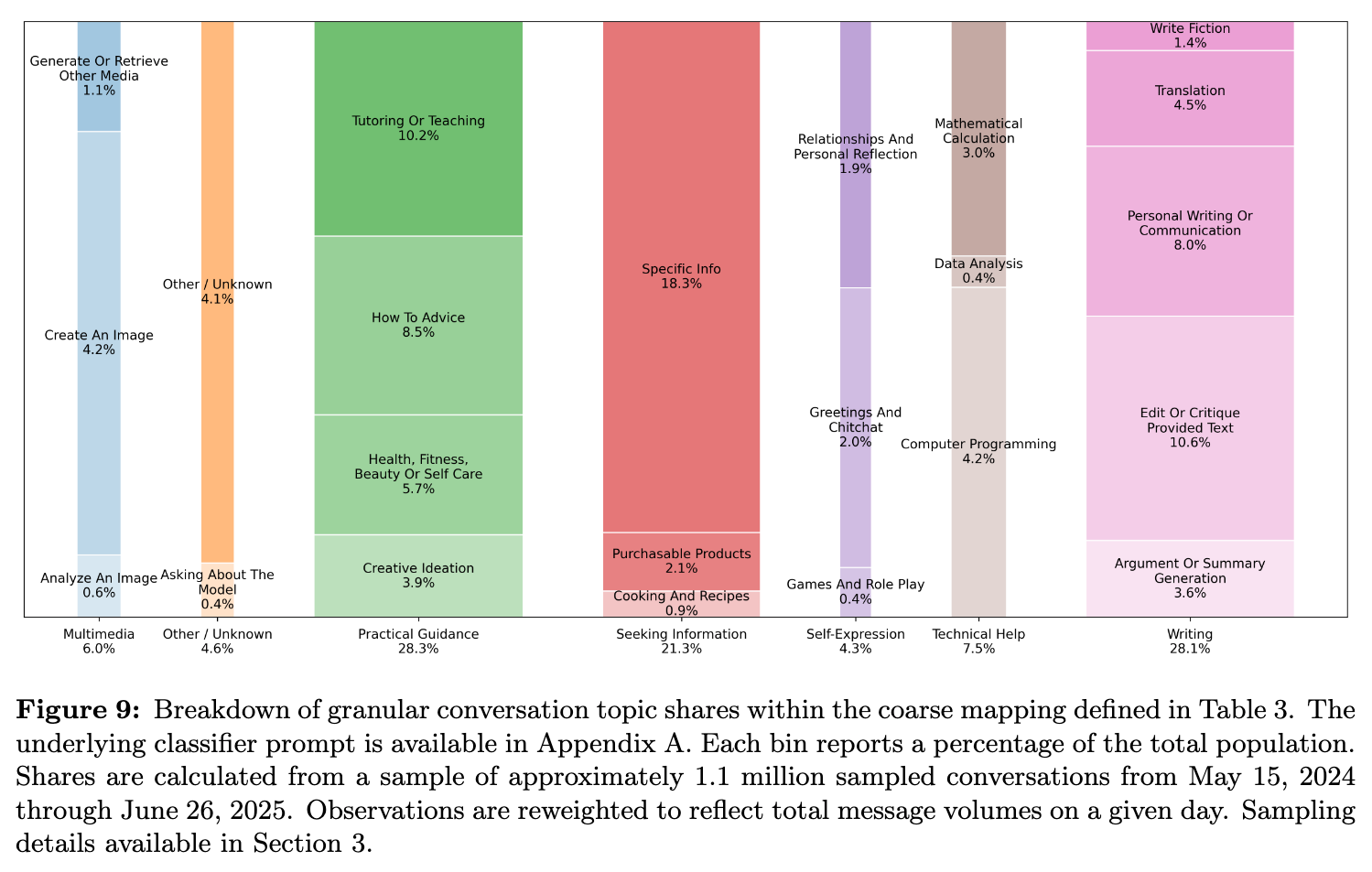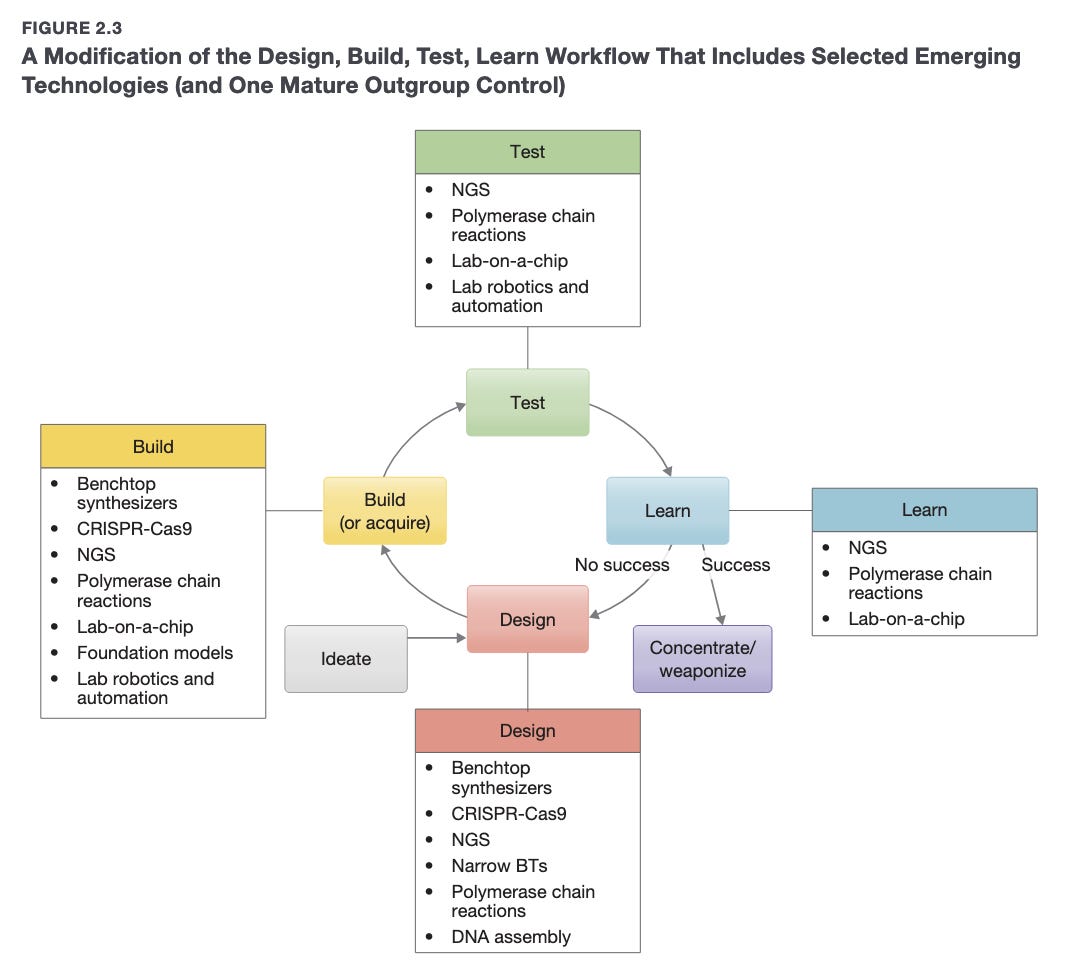
Happy Friday, colleagues. It’s the end of the week and once again I’m going through my long list of idle browser tabs trying to catch up where I can. Lots of R and AI-related news this week. Subscribe now Emil Hvitfeldt: Slidecrafting (slidecrafting-book.com) . This is a really wonderful one-stop shop for tips on making beautiful slides with reveal.js and Quarto.