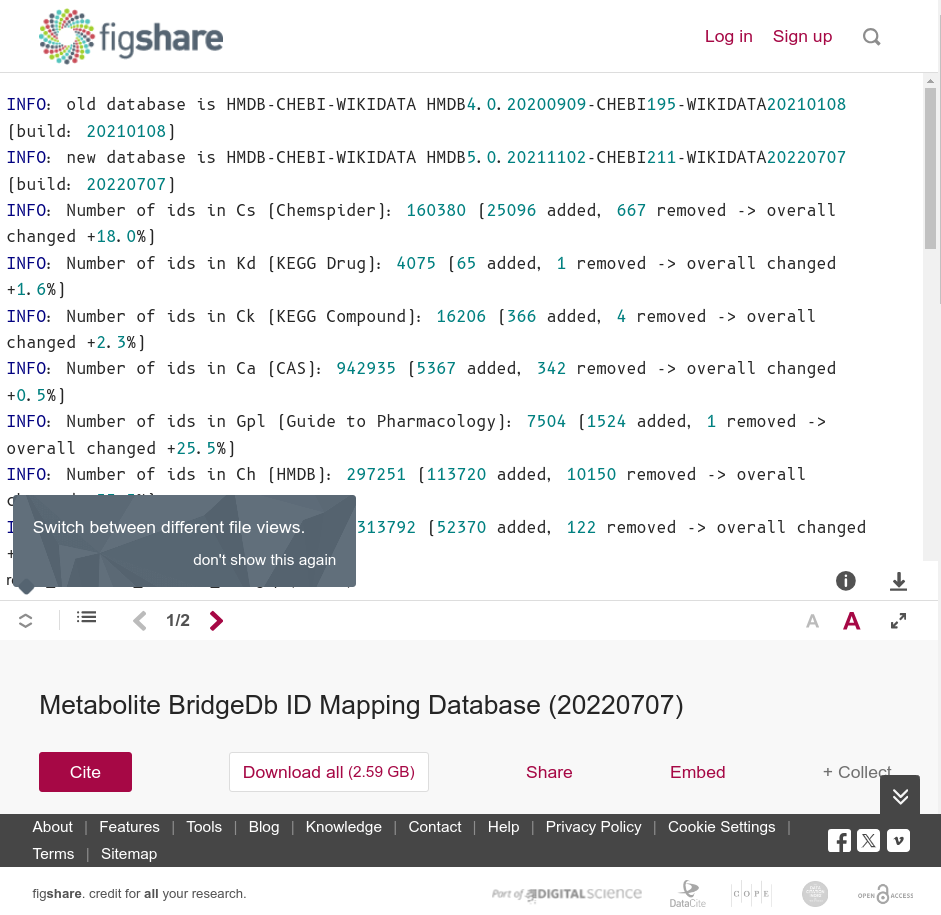
Open Science doesn’t make publishing easier. That that’s all for the better: our research efforts are complex, so why should the publishing be. Sure, I am not talking about references formatting or moving the Methods section to the right location, or some silly statement that all authors agree with the manuscript when you are the only author. No, let’s talk about data. What should you publish? How, and when?