
Somehow I get the feeling that botanists haven't got the "open data" religion. Not only is the list of plant names list behind a really bad license, but the Global Plants Initiative (GPI) hides its type images behind a JSTOR Plant Sciences paywall.

Somehow I get the feeling that botanists haven't got the "open data" religion. Not only is the list of plant names list behind a really bad license, but the Global Plants Initiative (GPI) hides its type images behind a JSTOR Plant Sciences paywall.

As part of the discussion on whether legacy biodiversity literature matters a graph from the following paper came up:So, why is the Sarkar et al. graph bogus? Here is their graph (Fig. 3) for animals:This is the number of new animal species described each year, estimated by parsing taxonomic names and extracting the date in the taxonomic authority. There are two prominent "spikes" which are worrying.

I've just come back from a pro-iBiosphere Workshop at Leiden where the role of "legacy literature" became the subject of some discussion.
Continuing the theme of trying to map specimens cited in the literature to the equivalent GBIF records, consider the GBIF record http://data.gbif.org/occurrences/685591320, which according to GBIF is specimen "ZFMK 188762" (a [sic] holotype of Praomys hartwigi ).This is odd, because the original publication of this name (Eisentraut, M. 1968 .Beitrag zur Saugetierfauna von Kamerun.
The rumour that Elsevier is buying Mendeley has been greeted with a mixture of horror, anger, peppered with a few congratulations, I told you so's, and touting for new customers:Here's some probably worthless speculation to add to the mix. Disclosure: I use Mendeley to manage 100,000's of references, and use the API for various projects.
Following on from my previous post bemoaning the lack of links between biodiversity data sets, it's worth looking at different ways we can build these links. Specifically, data can be tightly or loosely coupled. Tight coupling Tight coupling uses identifiers. A good example is bibliographic citation, where we state that one reference cites another by linking DOIs.
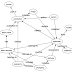
The journal Mycokeys has published the following paper:This paper contains a diagram that seems innocuous enough but which I find worrying:The nodes in the graph are "biodiversity megascience platforms", the edges are "cross-linkages and data exchange". What bothers me is that if you view biodiversity informatics through this lens then the relationships among these projects becomes the focus.

The Integrated Digitized Biocollections (iDigBio) project aims to advance digitising US biodiversity collections. They recently published a GUID Guide for Data Providers. In the PDF document I read this:My heart sank.

Say what you will about Elsevier, they are certainly exploring ways to re-imagine the scientific article. In a comment on an earlier post Fabian Schreiber pointed out that Elsevier have released an app to display phylogenies in articles they publish. The app is based on jsPhyloSVGand is described here.

Following on from the SVG experiments I've started to put some of the Javascript code for displaying phylogenies on Github. Not a repository yet, but as gists, little snippets of code. Mike Bostock has created http://bl.ocks.org/ which makes it possible to host gists as working examples, so you can play with the code "live".The first gist takes a Newick tree, parses it and displays a tree.
The following poem by David Maddison was published in Systematic Biology (doi:10.1093/sysbio/sys057) under a CC-BY-NC license.I think that I shall never seeA thing so awesome as the TreeThat links us all in paths of genesDown into depths of time unseen;Whose many branches spreading wideHouse wondrous creatures of the tide,Ocean deep and mountain tall,Darkened cave and waterfall.Among the branches we may findCreatures there of every