It’s a short week here in the US. As I reflect on the tools that shape modern bioinformatics and data science it’s striking to see how far we’ve come in the 20 years I’ve been in this field. Today’s ecosystem is rich with tools that make our work faster, better, enjoyable, and increasingly accessible.
Paired Ends
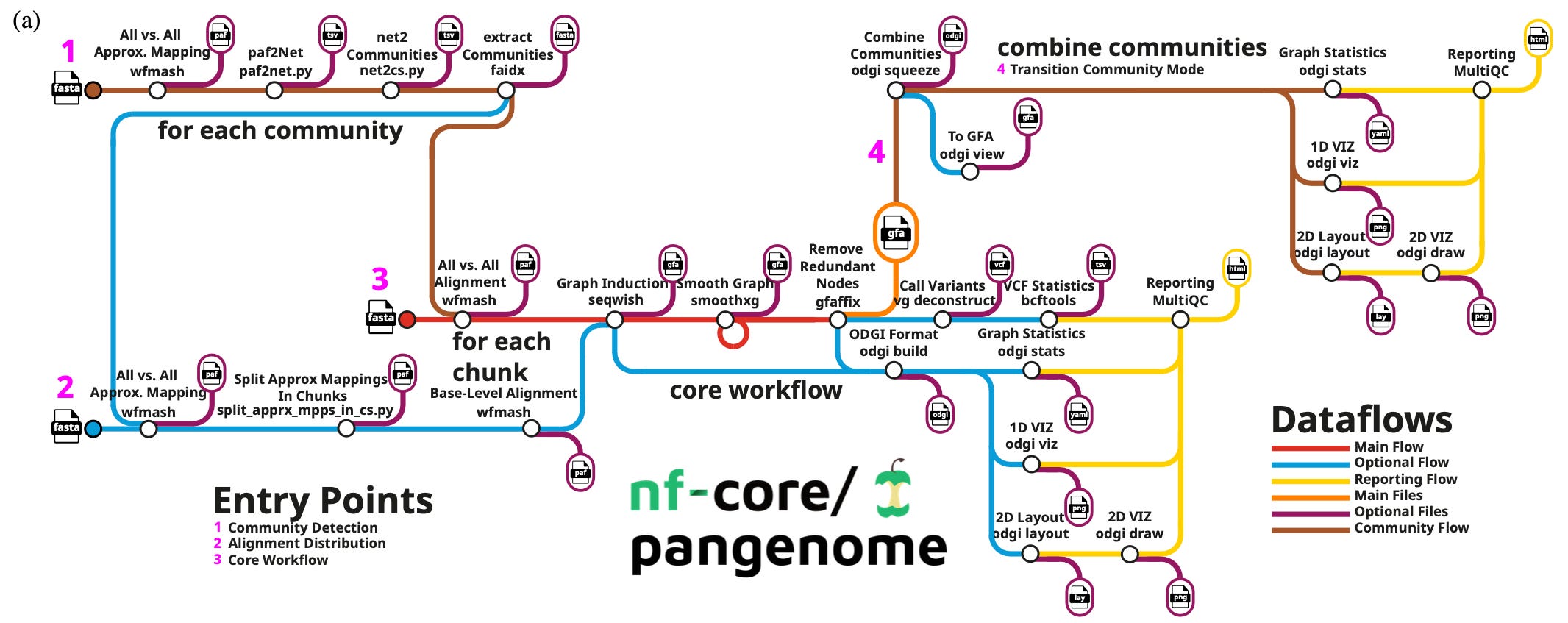
This week’s recap highlights pangenome graph construction with nf-core/pangenome, building pangenome graphs with PGGB, benchmarking algorithms for single-cell multi-omics prediction and integration, RNA foundation models, and a Nextflow pipeline for characterizing B cell receptor repertoires from non-targeted bulk RNA-seq data.

This post is inspired by the Bluesky Network Analyzer made by @theo.io. I’m encouraging everyone I know online to join the scientific community on Bluesky. In that post I link to several starter packs — lists of accounts posting about a topic that you can follow individually or all at once to start filling out your network. I started following accounts of people I knew from X and from a few starter packs I came across.

I joined Twitter1 way back in 2009. For nearly 10 years “scitwitter” was an amazing place for discussion, discovery, and engagement with the scientific community. The #Rstats and #pydata hashtags were great places to learn about something new in programming, #icanhazpdf was great for getting papers you didn’t have access to, and conference live-tweeting was common and useful for those of us with FOMO not able to make it in person.

This week’s recap highlights an AI agent for automated multi-omic analysis (AutoBA), rapid species-level metagenome profiling and containment (sylph), a review on genome-wide association analysis beyond SNPs, private information leakage from scRNA-seq count matrices, and a method to “unlearn” viral knowledge in protein language models as a means to develop safe PLM-based variant effect analysis (PROEDIT). Others that caught my attention include
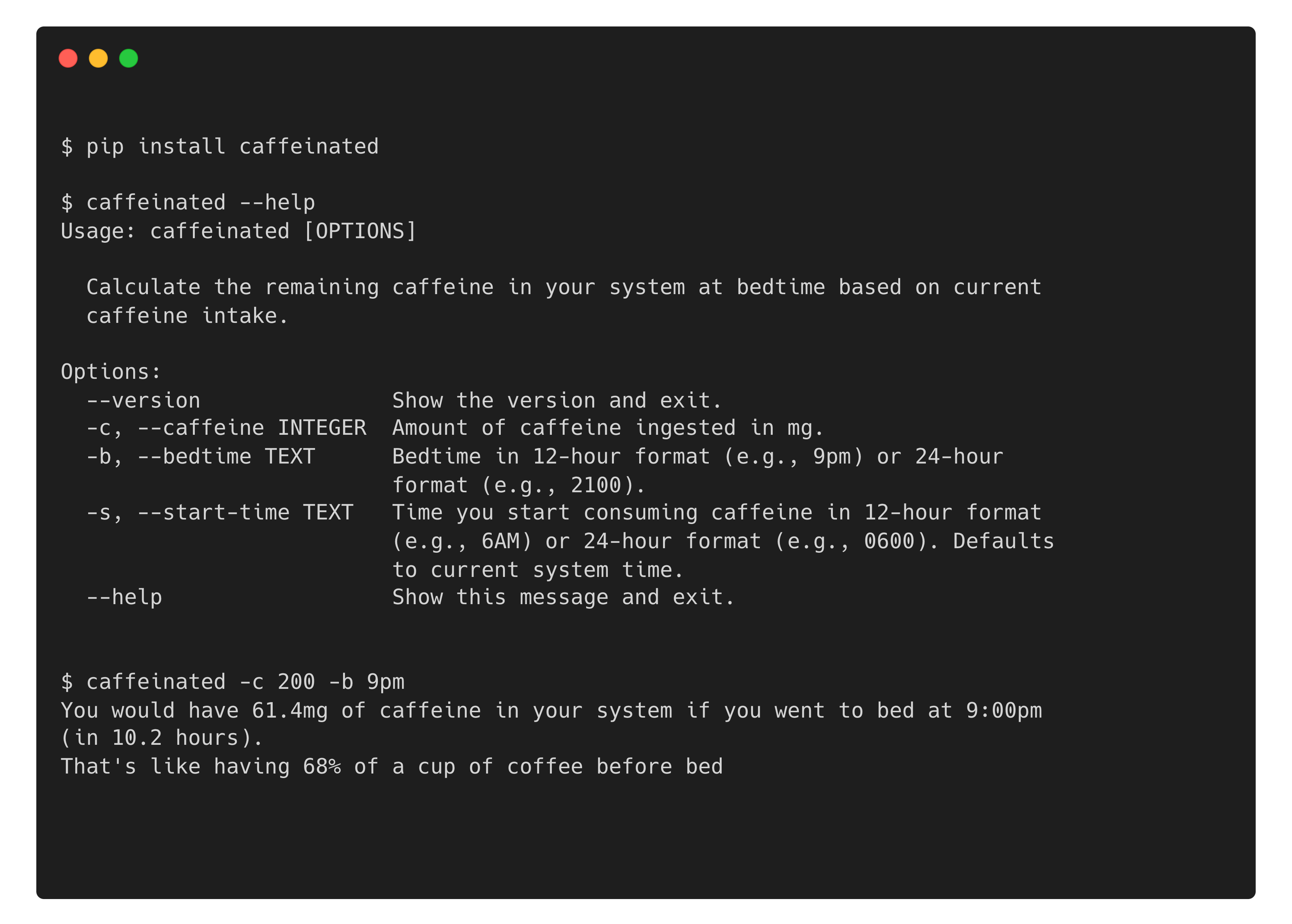
In the spirit of Learning in Public, I wanted an excuse to explore (1) click for creating command line interfaces, (2) Cookiecutter project templates, and (3) modern tools in the Python packaging ecosystem. If you’re primarily an R developer like me, I recently wrote about resources for getting better at Python for R users.
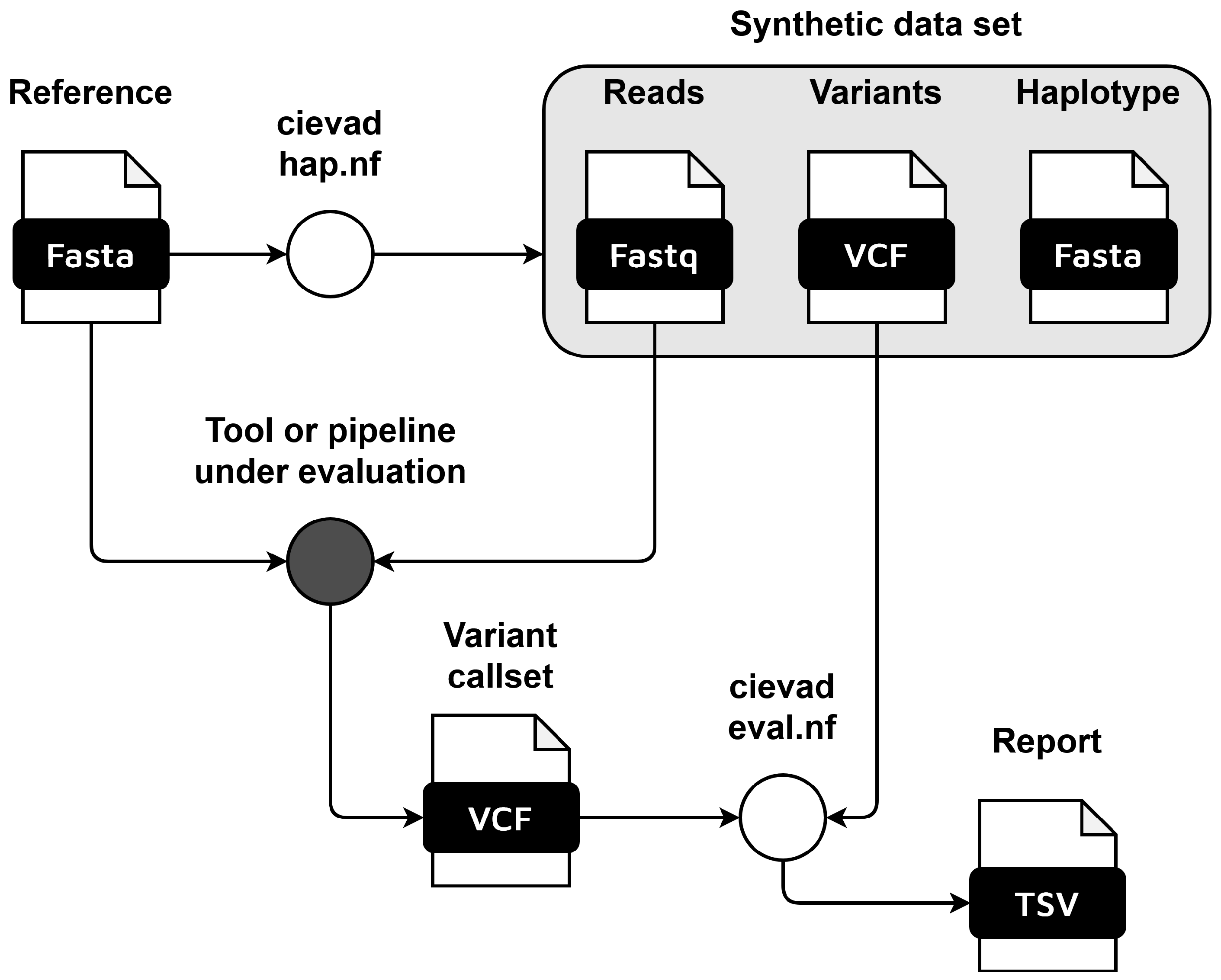
This week's recap highlights a new pipeline for metagenome quality assessment and taxonomic annotation (MAGFlow &

I just returned from a week in Barcelona where I attended the Nextflow Summit and nf-core hackathon, and I can hardly contain my excitement for the near term future of bioinformatics, computational biology, and open science in general.
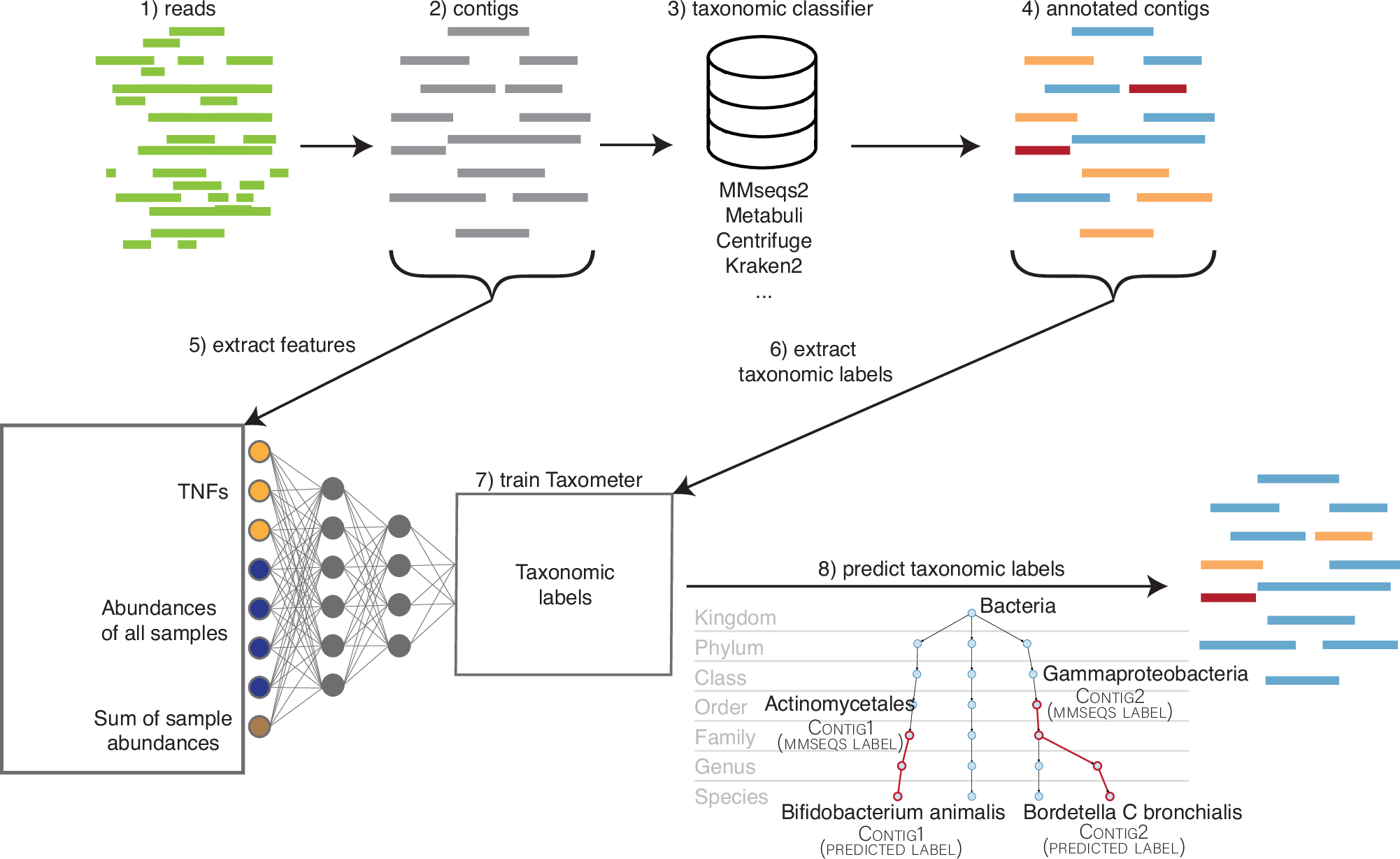
This week’s recap highlights protein design with RoseTTAFold, surveillance with wastewater sequencing, T2T human genomes, Vitessce for visualization of multimodal spatial single-cell data, and Taxometer for taxonomic classification of metagenomics contigs.

A Google search for “R vs Python” returns thousands of hits across sites like Reddit, IBM, Datacamp, Coursera, Kaggle, and many others. A quick Google Trends analysis shows that this search query has grown steadily over the last decade. Any real data scientist would agree that this argument is silly, that the right answer is to use the best tool for the job. What’s “best” isn’t always easy to answer.

This week’s recap highlights a new Nextflow workflow for calculating polygenic scores with adjustments for genetic ancestry, a paper demonstrating that whole exome plus imputation on more samples is more powerful than whole genome sequencing for finding more trait associated variants, a new deep-learning-based splice site predictor that improves spliced alignments, a new method for accurate community profiling of large metagenomic datasets, and