
posit::conf(2025) & Nextflow Summit talks, bioRxiv+medRxiv AI reviews, R updates (R Data Scientist, R weekly), Claude Scientific Skills, Python package development for R users, new papers &

posit::conf(2025) & Nextflow Summit talks, bioRxiv+medRxiv AI reviews, R updates (R Data Scientist, R weekly), Claude Scientific Skills, Python package development for R users, new papers &

29 talks from the 2025 Nextflow Summit are now available on YouTube

Over 100 recorded talks from posit::conf(2025) are now available on YouTube
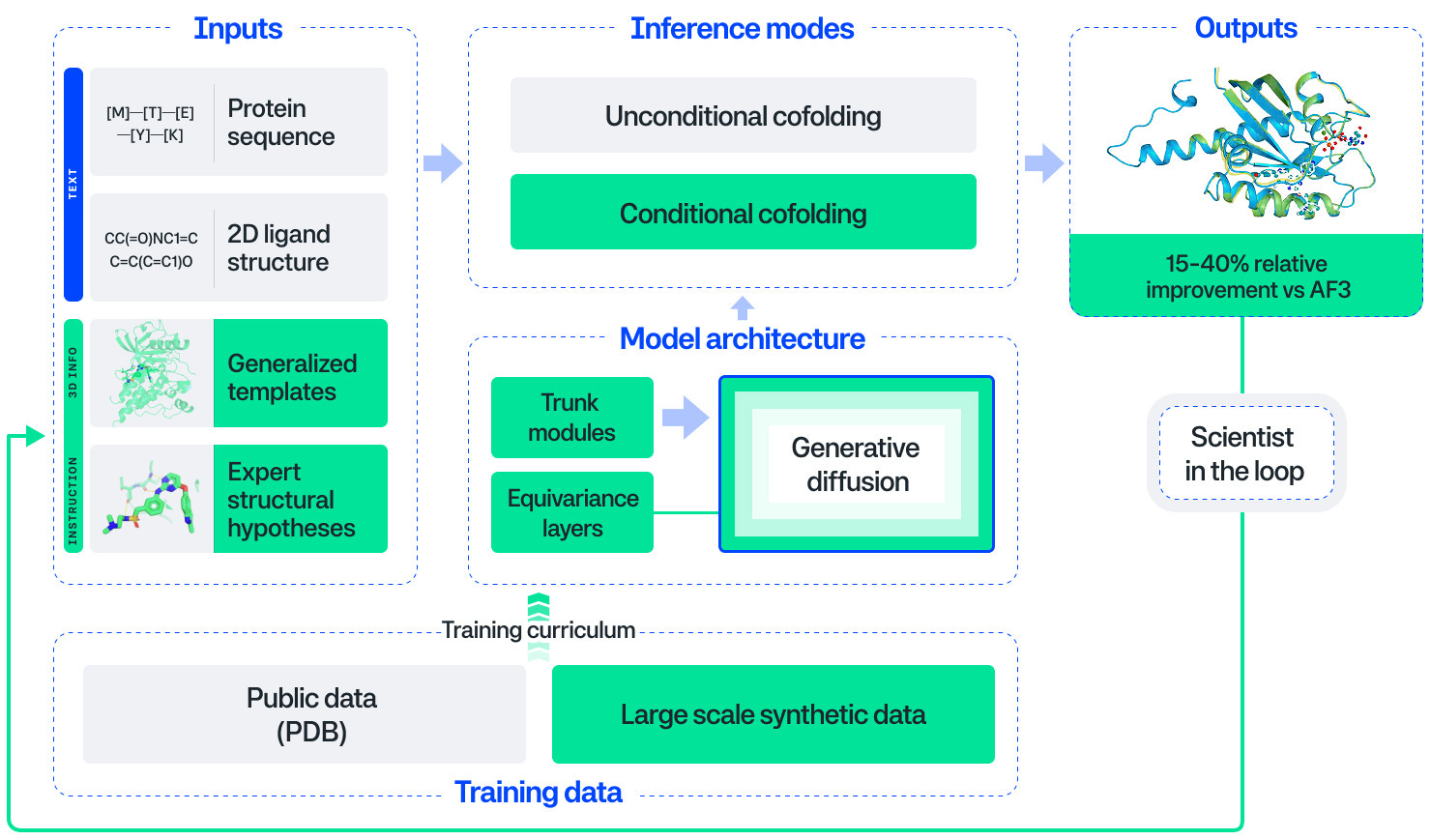
LLMs for learning to code, R updates (R Data Scientist, R Weekly), RAG, layoffs & AI, DuckDB for data science, Anthropic+Iceland, AI in science, AI in drug discovery, Nextflow &

WIRED published a special issue with 17 readings from the furthest reaches of the AI age

59 new software packages, updates for genomics, single-cell transcriptomics, spatial omics, QC tooling, core I/O, GFF/CIGAR handling, much more.

AI in science, Quarto, biodiversity, changed grant titles, new papers &
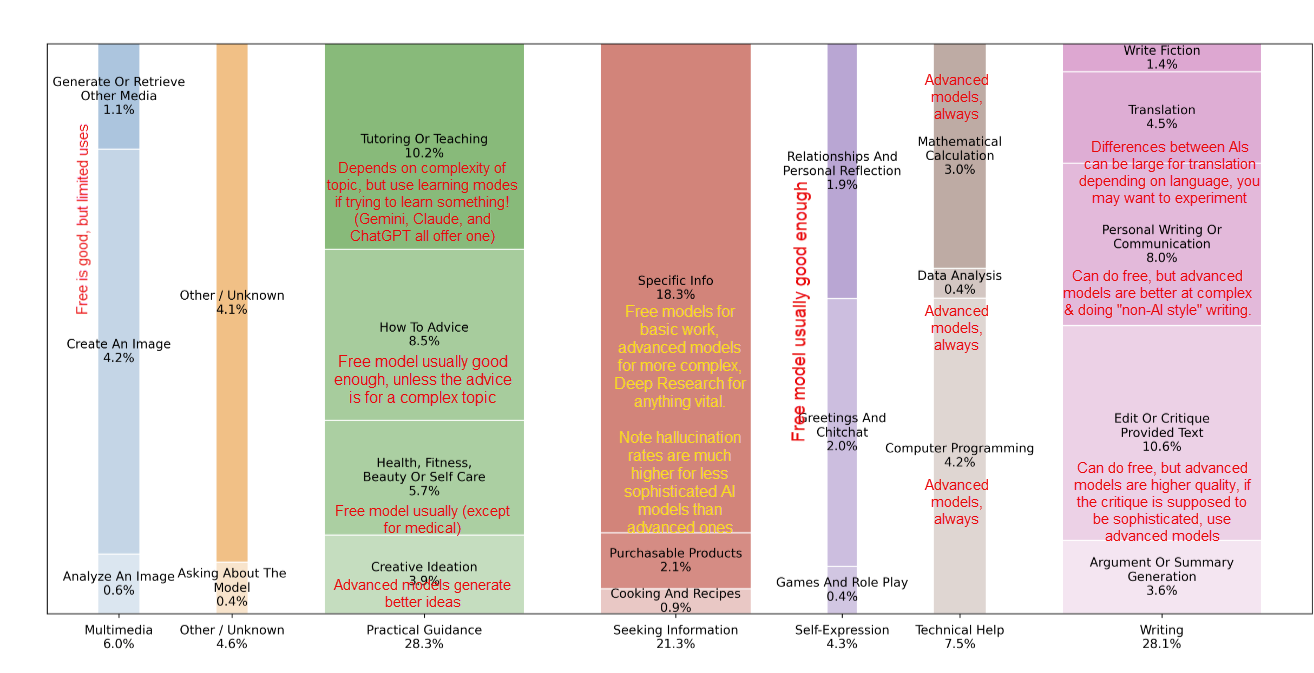
Polyglot data science pipeline (R+Python+Julia) with rixpress, EuroBioC2025/BCN, Python packaging for R devs, Python in 2025, R/Pharma, DARPA, ASHG, the R Data Scientist, Quarto, universities and AI
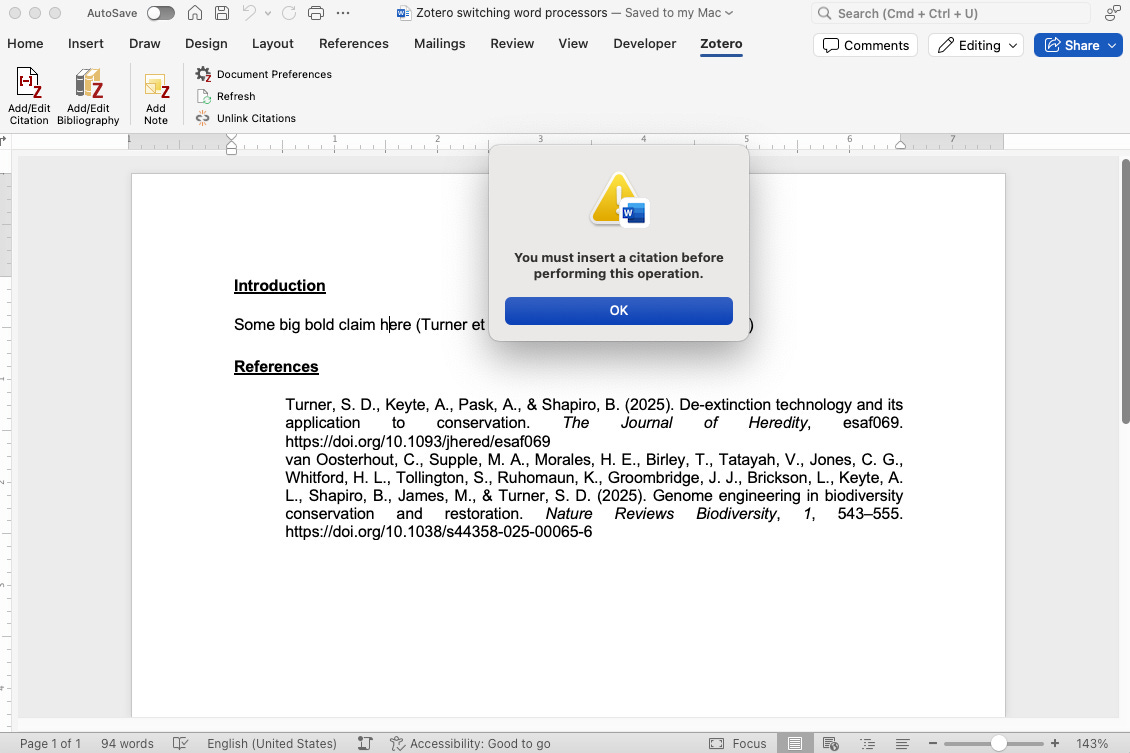
Moving from Google Docs to Word will break your Zotero citations/bibliography linkages unless you do this first.
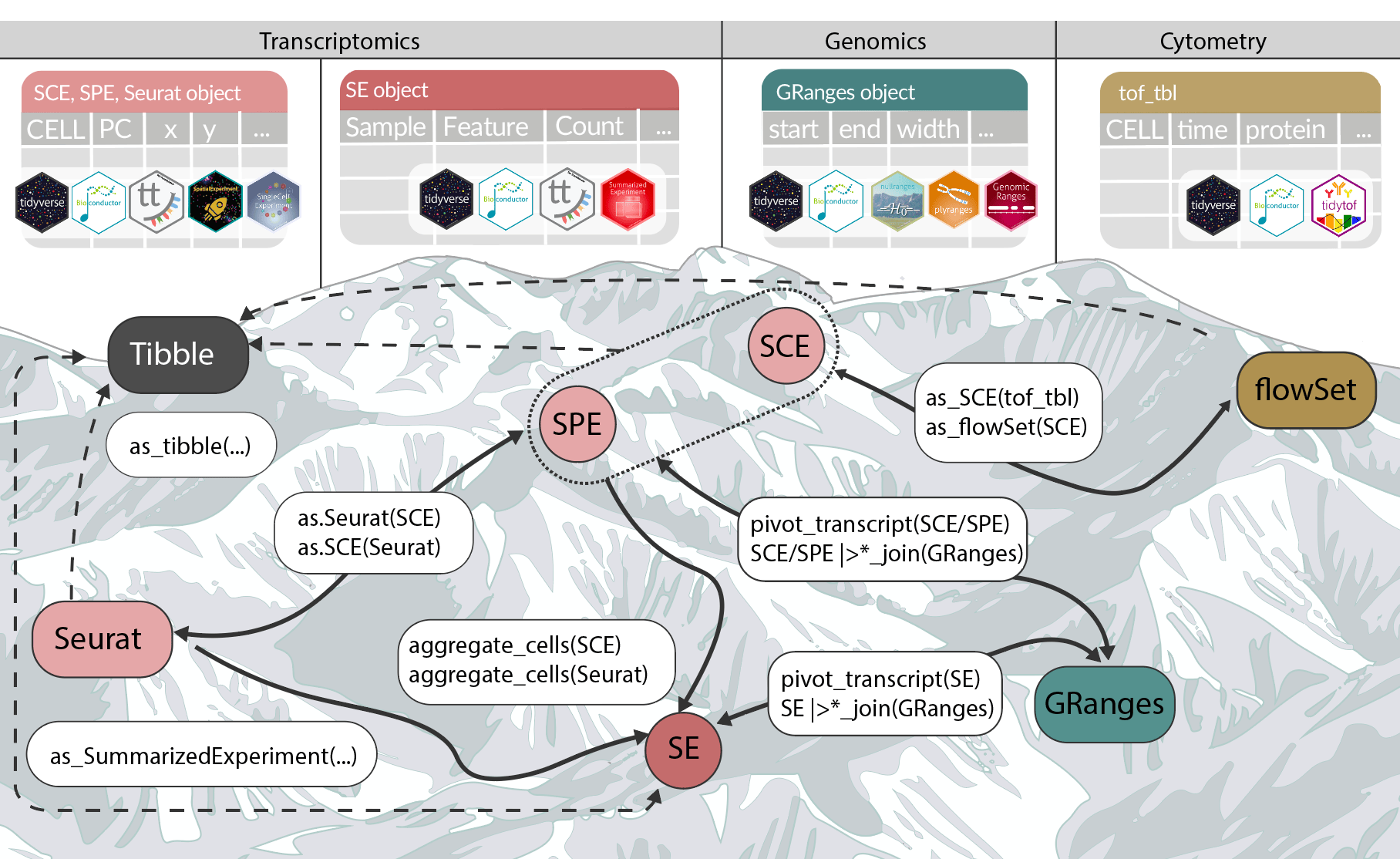
Bioconductor on GPUs, tidyomics, Quarto 1.8, Anthropic red-teaming on AIxBio, PGS in the clinic, Shock Doctrine in genome engineering, GenAI for data viz with R, Datapalooza, 4 compact rejections
I recommend subscribing to Claus Wilke’s newsletter, Genes, Minds, Machines. I’ve linked to many of his essays in recent weeks. This one was a good read. Now that I’m back in academia and will be taking on Ph.D. students in my lab, this was good advice for me to read, as a future mentor.