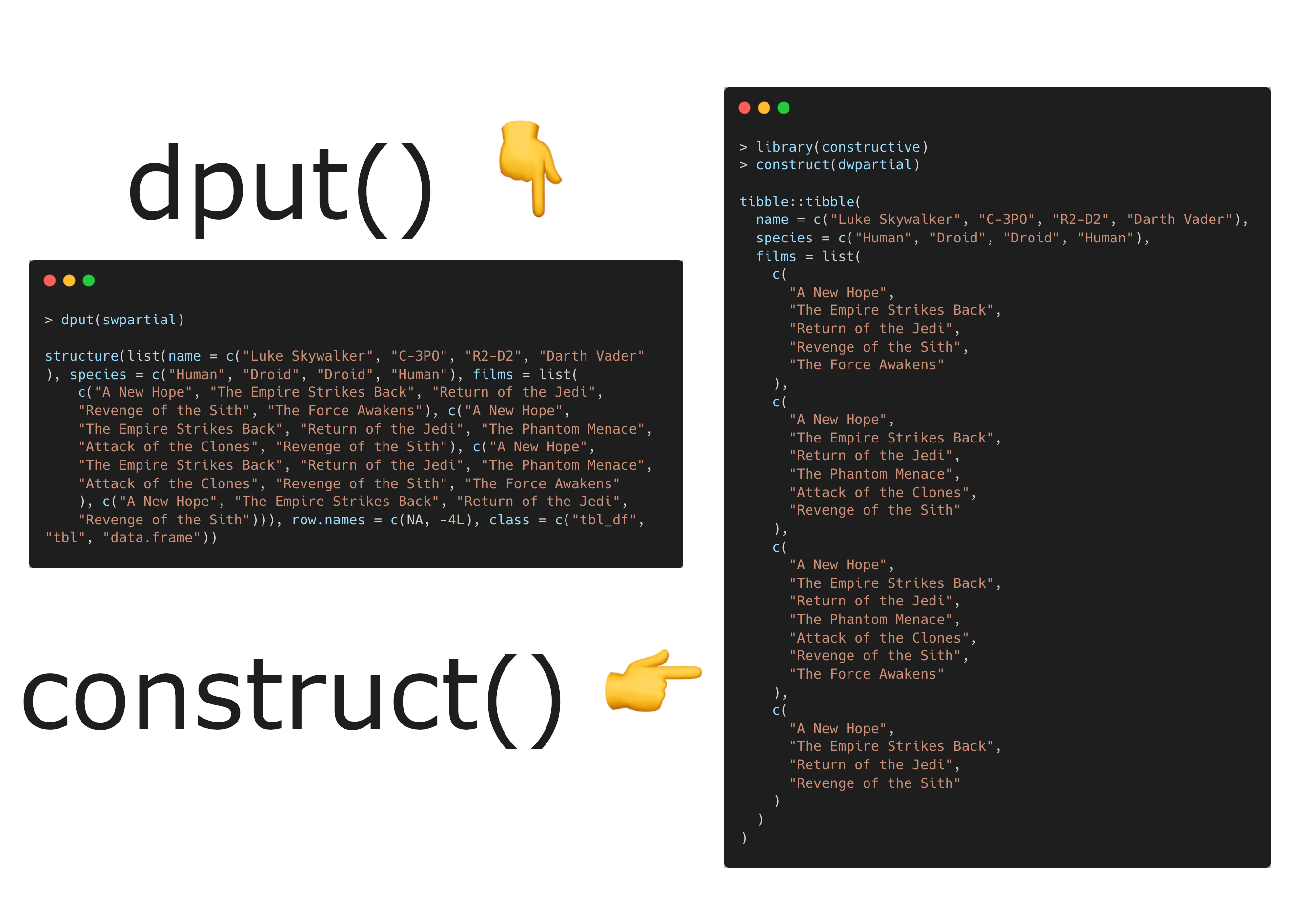
Today I discovered the constructive package and the construct() function for creating R objects with idiomatic R code to make human-readable reproducible examples. CRAN: https://cran.r-project.org/package=constructive Source: https://github.com/cynkra/constructive/ Docs &