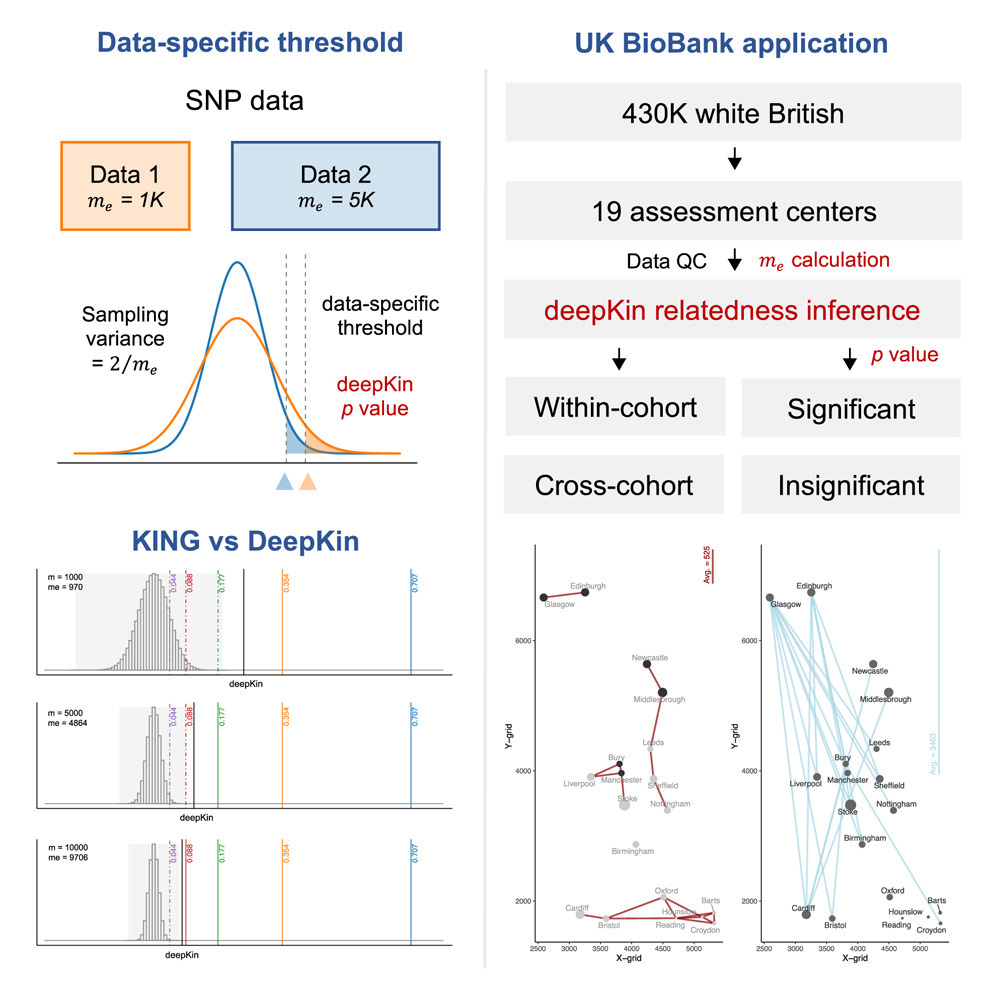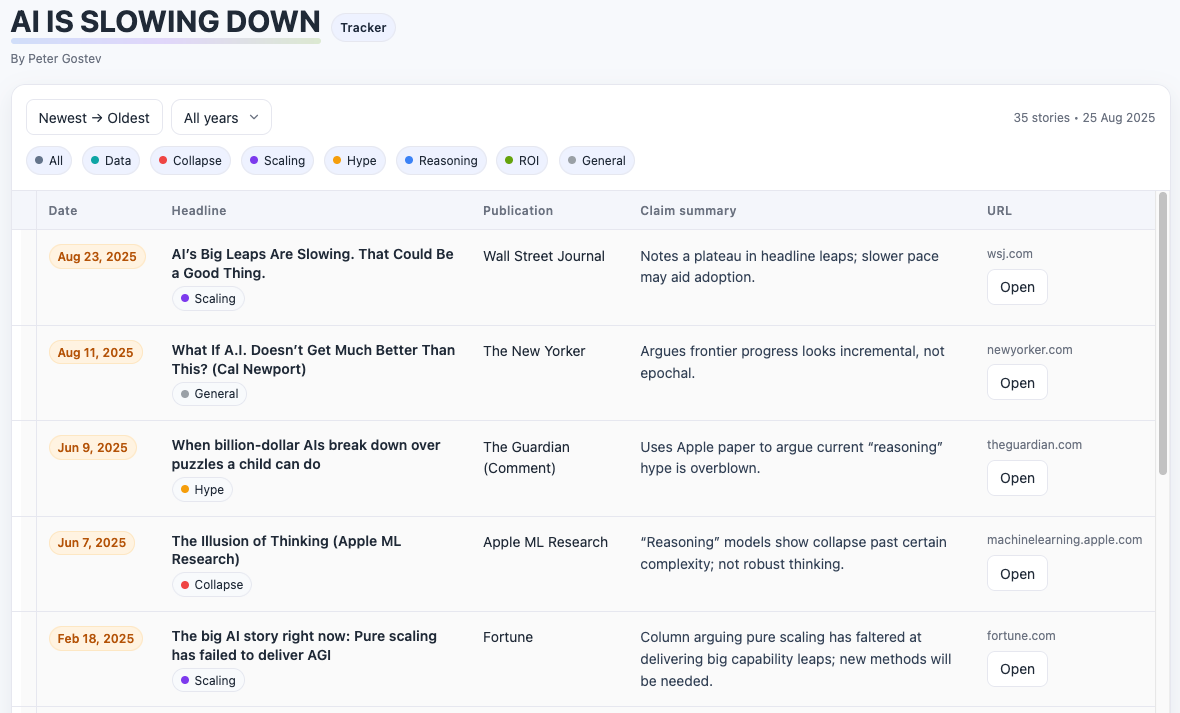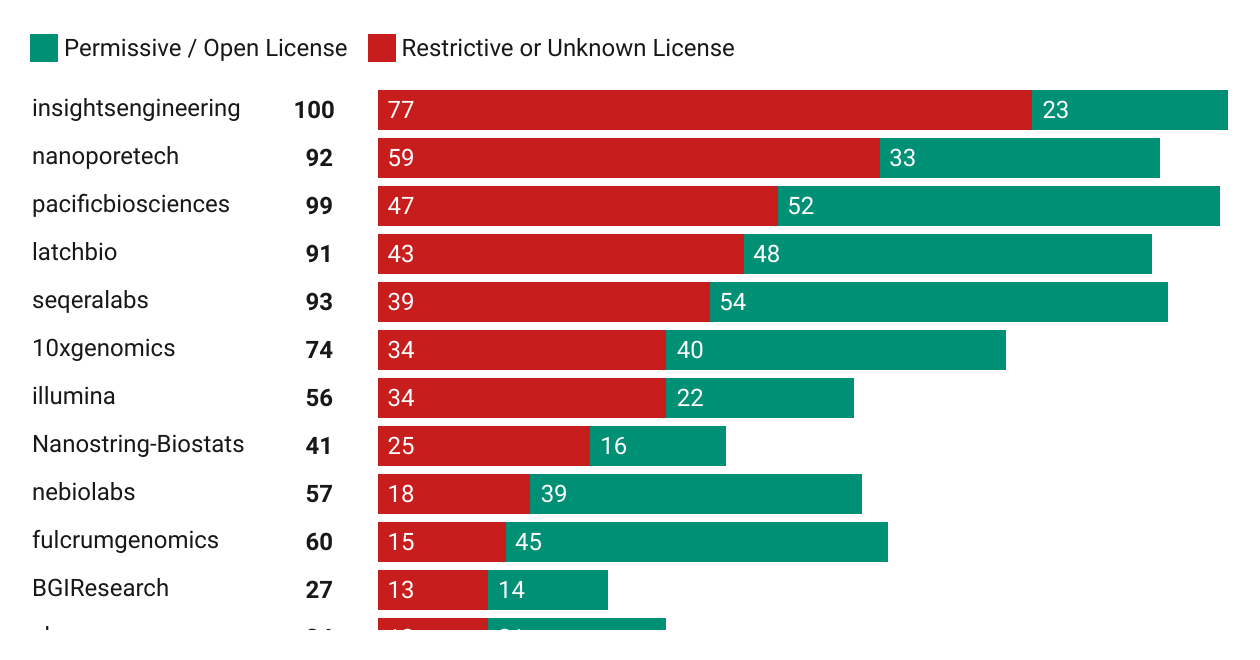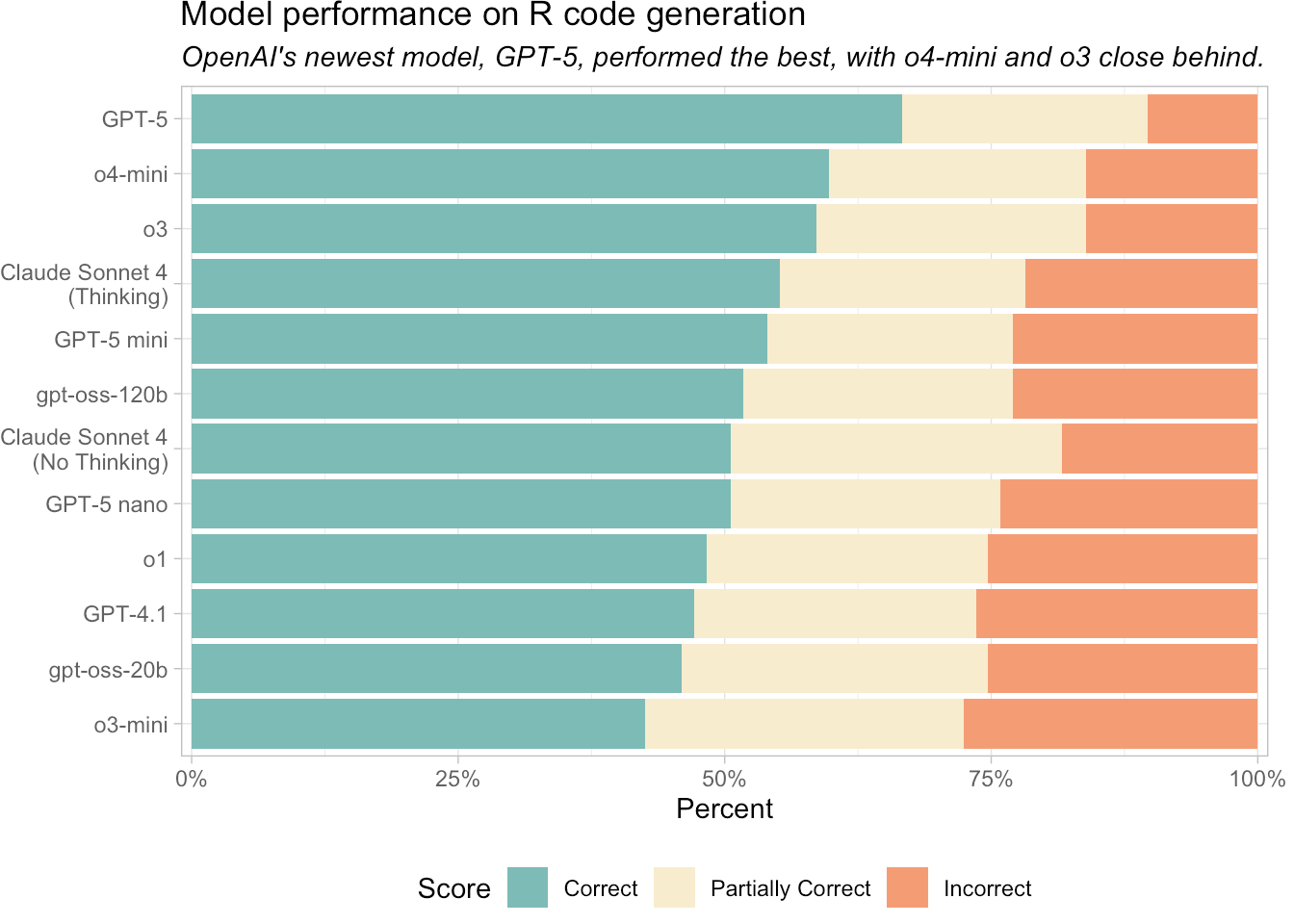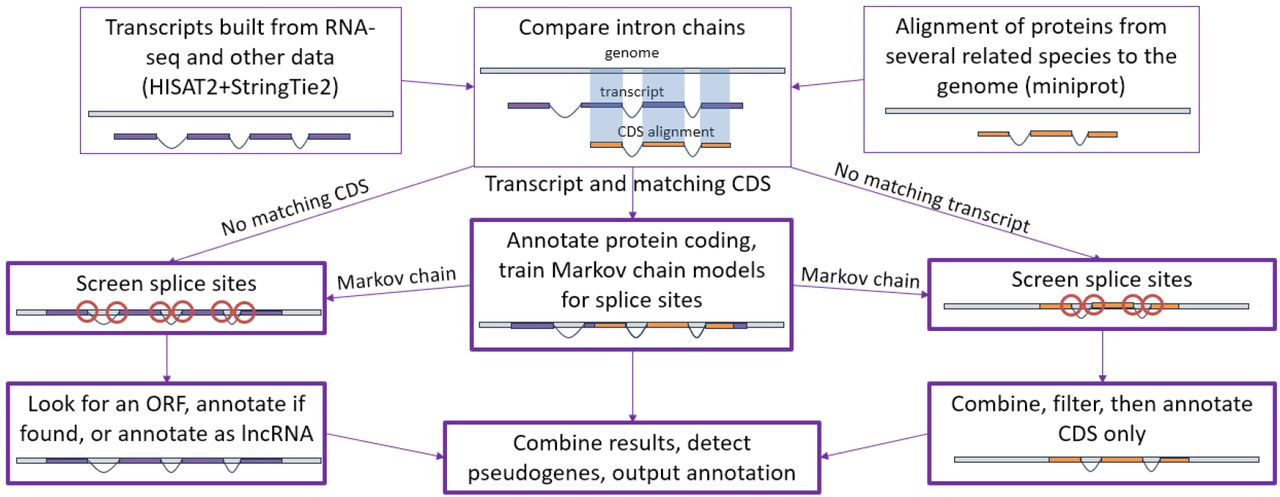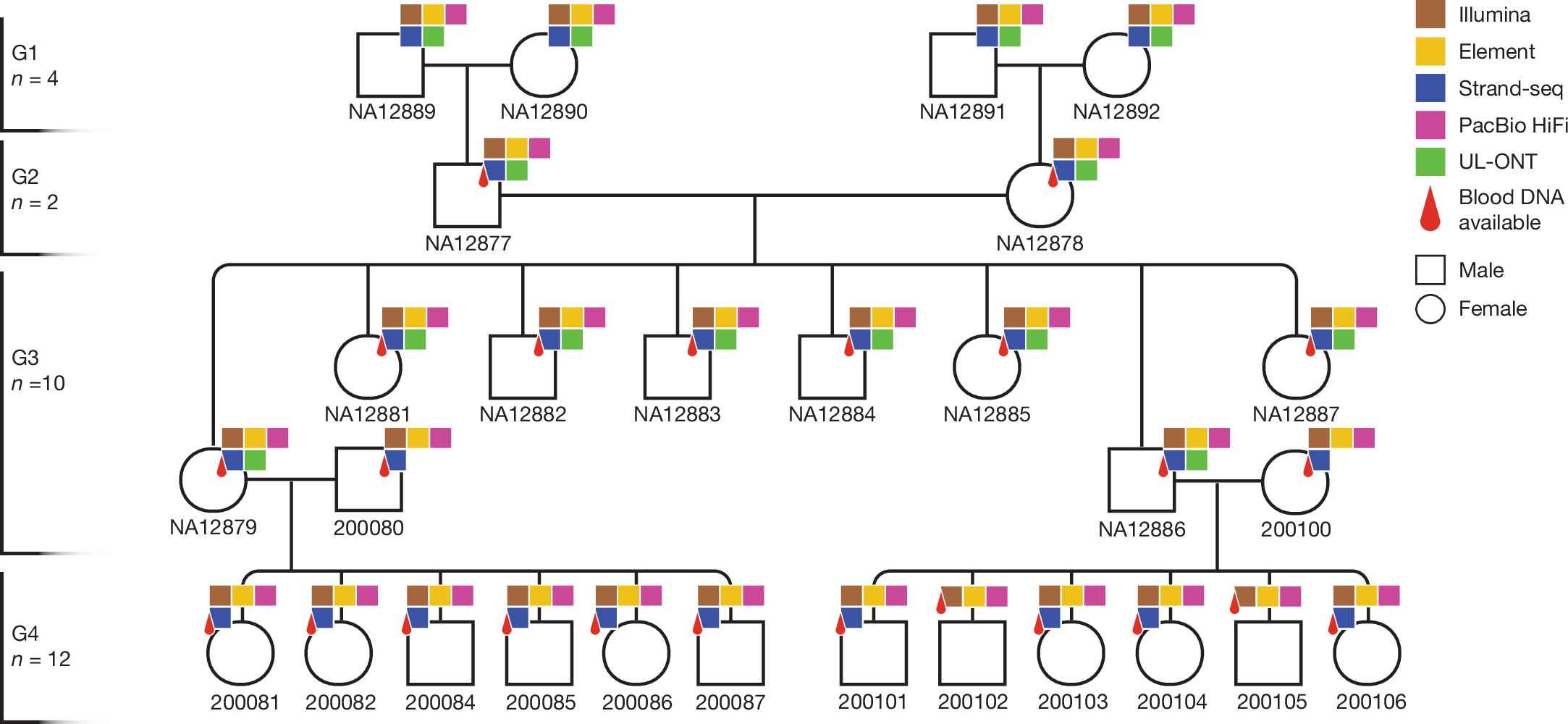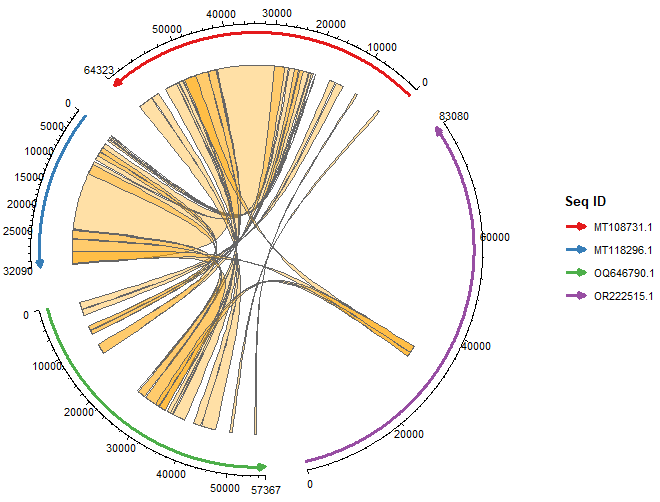
Happy Friday, colleagues. Somehow it’s September (I did not approve of this). Lots going on this week, and this is my regular attempt to close out my browser tabs I’ve accumulated over the past week with blog posts, podcasts, papers, etc. in AI, data science, genomics, public health, programming, scicomm, and other miscellany.