I missed this paper (Yuan et al. 2024) when it came out last year, but my friend and colleague Jeremiah Scott brought it to my attention. The bit on nuchal sesamoids in shrews is so good and so weird that I’m just going to copy and paste it in its entirety.
Rogue Scholar Posts
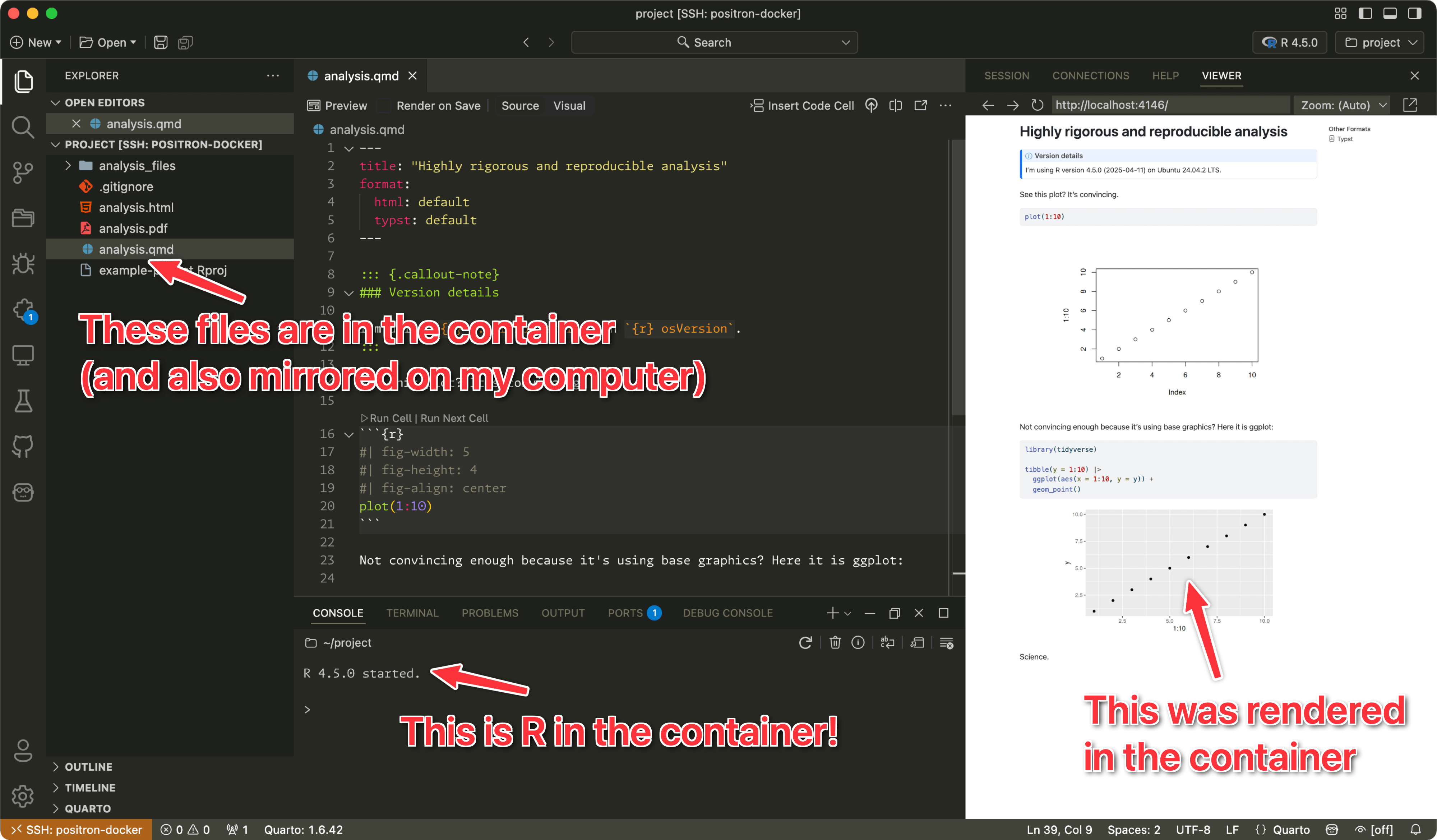
I’ve long been a proponent of making quantitative research reproducible. It’s the main reason I do all my scientific writing in Quarto—I can mix code and text in the same document so I don’t need to copy/paste numbers, tables, and figures from some statistical program into a word processor. Everything automatically ends up one compiled document based on the most current data.
(This was buried in Part 5 of my 2011 review of the Sideshow Apatosaurus maquette, but it’s long deserved to be a post of its own, and now it is. I’m not adding anything new here, just extracting and reposting the relevant bits, for reasons that will become clear in a future post.

My blog focuses on the two primary ways "AI"—or more accurately, transformer-based models—are impacting academic search.
KI-Agenten erweitern die Möglichkeiten von KI-Systemen erheblich, doch die rechtlichen Rahmenbedingungen dafür sind noch im Aufbau. Dadurch ergeben sich neue Fragen zu Verantwortung und Kontrolle. „2025 ist das Jahr der KI-Agenten“, prognostizierte Kevin Weil, Chief Product Officer von OpenAI, Anfang Januar beim Weltwirtschaftsforum in Davos.

Edward G Robinson’s gangster Johnny Rocco in Key Largo (John Huston, 1948) seems uncertain when asked by Humphrey Bogart and Lionel Barrymore’s sequestered characters what he wants. But after some prompting, he comes up with ‘That’s it. I want more.’ Mindless acquisitiveness matches his mendacity and violence. And that’s the story with the discreditable alliance between pro sports and the screen.

Nous vous proposons en cette fin d’année universitaire une nouvelle édition de notre bulletin de veille, particulièrement riche en trucs et astuces. Les fonctionnalités et modules ne sont pas en reste, avec notamment la publication de l’application Zotero pour Android.

This week’s recap highlights the Rust-based wgatools for manipulating alignments and visualizing in the terminal, the nf-core scnanoseq Nextflow pipeline for ONT scRNA-seq, sawfish for better SV discovery and genotyping with long reads, the BINSEQ high-performance binary formats for nucleotide sequence data, and a unified analysis of atlas single-cell data.
The National Institute for Fusion Science (NIFS) is an inter-university research institute based in Japan. NIFS promotes academic collaborative research and also strives to meet the expectations of the industrial sector, which is beginning to engage with fusion energy development, as well as students, including those pursuing recurrent education.
I say “semi-spoileriffic” because I’m not going to go out of my way to give away any plot points or creature details you couldn’t get from watching the trailers and TV spots, but if you want to keep yourself pure as the driven snow, you might want to save this post for later.
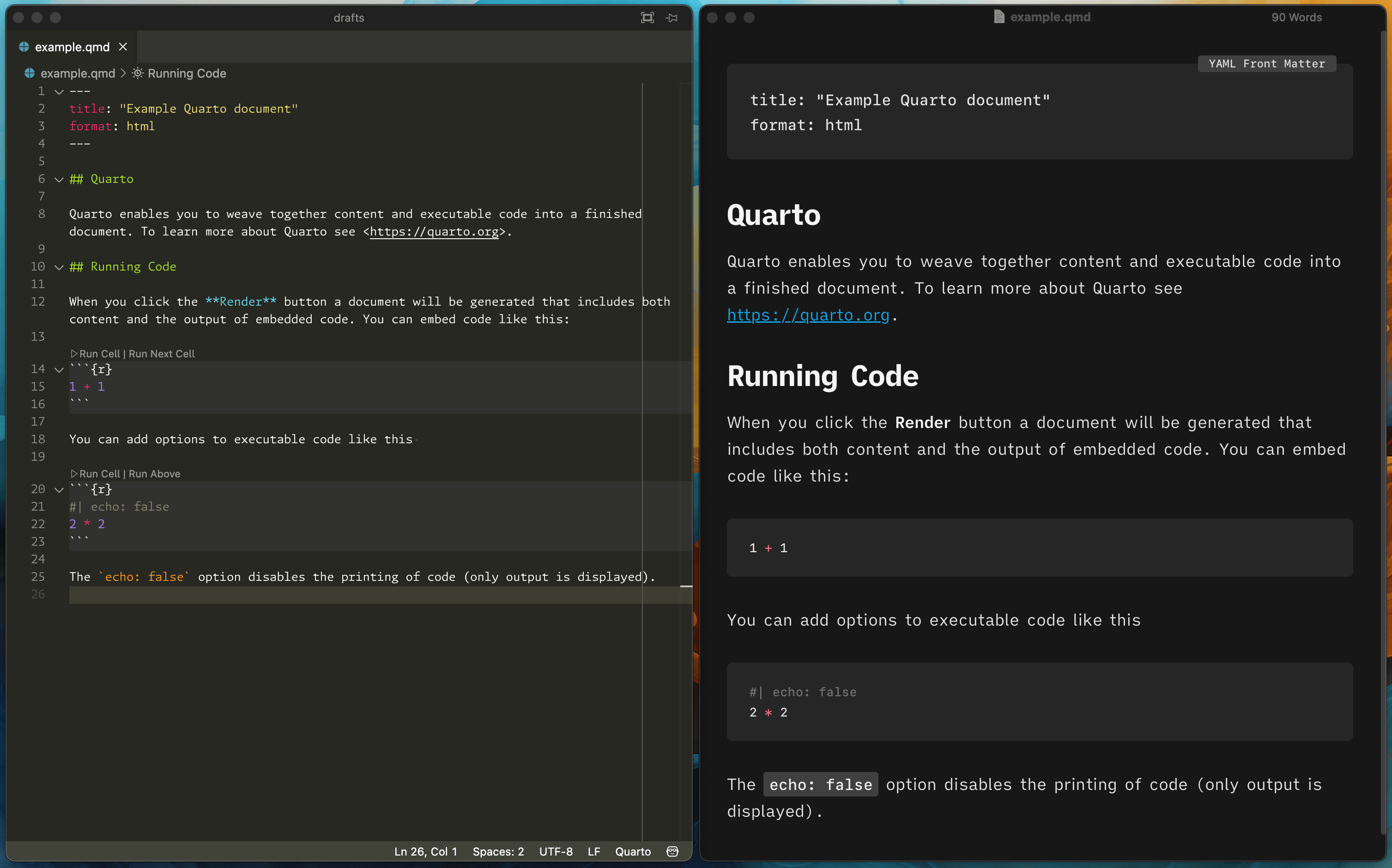
Positron’s Quarto support is really quite robust and it’s full of nice little features like previewing a citation using whatever bibliography style your document is using (Chicago, APA, etc.) when you hover over a citation key: …or knowing about the different headings inside your document for tab-completed cross references: …or even knowing about those headings in other documents in a project: That’s all neat, but for long-form writing, like